
Micrococcus terreus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Micrococcus
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
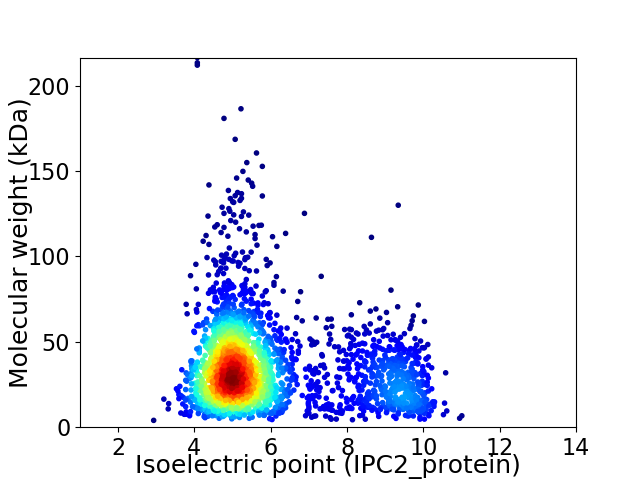
Virtual 2D-PAGE plot for 2759 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
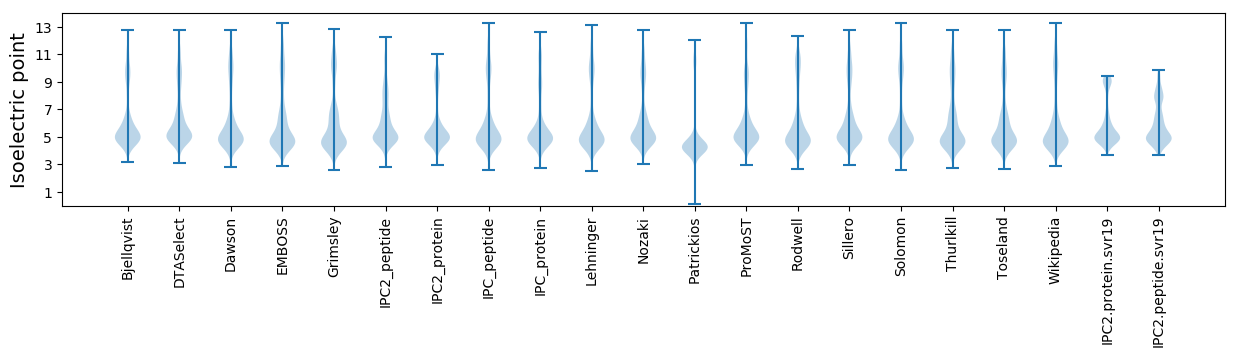
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I7ML73|A0A1I7ML73_9MICC Nucleoside-diphosphate-sugar epimerase OS=Micrococcus terreus OX=574650 GN=SAMN04487966_104247 PE=4 SV=1
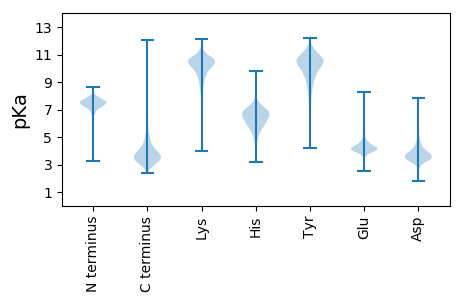
MM1 pKa = 6.63TTAHH5 pKa = 7.64CIRR8 pKa = 11.84HH9 pKa = 5.15TMTALGAVTALILMTGCADD28 pKa = 3.27QFRR31 pKa = 11.84AIDD34 pKa = 3.72SADD37 pKa = 3.52SEE39 pKa = 4.92GPSAHH44 pKa = 6.7GAQSSTNEE52 pKa = 3.95SRR54 pKa = 11.84LEE56 pKa = 4.08TIKK59 pKa = 10.33TSGVLNACTTGDD71 pKa = 3.85YY72 pKa = 11.12PPFTEE77 pKa = 4.29DD78 pKa = 3.11TGDD81 pKa = 3.83GEE83 pKa = 4.51LTGIDD88 pKa = 3.21VDD90 pKa = 4.1MAADD94 pKa = 3.58LAEE97 pKa = 4.11TLEE100 pKa = 4.83VDD102 pKa = 4.33IEE104 pKa = 4.08WHH106 pKa = 4.36QTSWGDD112 pKa = 4.47LMDD115 pKa = 4.67TFLSDD120 pKa = 3.47CDD122 pKa = 3.48IAVGGISMNPPRR134 pKa = 11.84AEE136 pKa = 3.51QVFFSDD142 pKa = 6.18PILEE146 pKa = 4.63DD147 pKa = 3.27GKK149 pKa = 9.63TSIARR154 pKa = 11.84CDD156 pKa = 3.41EE157 pKa = 4.05VQNYY161 pKa = 6.89QTIEE165 pKa = 4.96DD166 pKa = 3.8INQPNVTSIFPEE178 pKa = 4.98GGTNEE183 pKa = 3.89AFARR187 pKa = 11.84EE188 pKa = 4.23HH189 pKa = 6.49FPNGDD194 pKa = 3.68LKK196 pKa = 10.18THH198 pKa = 7.12DD199 pKa = 4.13NLTIFDD205 pKa = 4.08EE206 pKa = 4.73LVAGGADD213 pKa = 3.49VMTTDD218 pKa = 3.77RR219 pKa = 11.84AEE221 pKa = 4.14VLYY224 pKa = 10.24IAHH227 pKa = 7.25EE228 pKa = 4.02YY229 pKa = 11.19DD230 pKa = 3.58EE231 pKa = 5.13LCAVNPDD238 pKa = 3.22EE239 pKa = 5.1TYY241 pKa = 10.95DD242 pKa = 3.93FSVMGYY248 pKa = 7.76MLPQGDD254 pKa = 4.43VVFKK258 pKa = 10.6EE259 pKa = 5.38YY260 pKa = 10.92VDD262 pKa = 3.4QWLTIALKK270 pKa = 10.7DD271 pKa = 3.53GTYY274 pKa = 11.05DD275 pKa = 3.89EE276 pKa = 5.35IAEE279 pKa = 4.11PWVGNVEE286 pKa = 5.61LIPEE290 pKa = 4.37DD291 pKa = 3.36
MM1 pKa = 6.63TTAHH5 pKa = 7.64CIRR8 pKa = 11.84HH9 pKa = 5.15TMTALGAVTALILMTGCADD28 pKa = 3.27QFRR31 pKa = 11.84AIDD34 pKa = 3.72SADD37 pKa = 3.52SEE39 pKa = 4.92GPSAHH44 pKa = 6.7GAQSSTNEE52 pKa = 3.95SRR54 pKa = 11.84LEE56 pKa = 4.08TIKK59 pKa = 10.33TSGVLNACTTGDD71 pKa = 3.85YY72 pKa = 11.12PPFTEE77 pKa = 4.29DD78 pKa = 3.11TGDD81 pKa = 3.83GEE83 pKa = 4.51LTGIDD88 pKa = 3.21VDD90 pKa = 4.1MAADD94 pKa = 3.58LAEE97 pKa = 4.11TLEE100 pKa = 4.83VDD102 pKa = 4.33IEE104 pKa = 4.08WHH106 pKa = 4.36QTSWGDD112 pKa = 4.47LMDD115 pKa = 4.67TFLSDD120 pKa = 3.47CDD122 pKa = 3.48IAVGGISMNPPRR134 pKa = 11.84AEE136 pKa = 3.51QVFFSDD142 pKa = 6.18PILEE146 pKa = 4.63DD147 pKa = 3.27GKK149 pKa = 9.63TSIARR154 pKa = 11.84CDD156 pKa = 3.41EE157 pKa = 4.05VQNYY161 pKa = 6.89QTIEE165 pKa = 4.96DD166 pKa = 3.8INQPNVTSIFPEE178 pKa = 4.98GGTNEE183 pKa = 3.89AFARR187 pKa = 11.84EE188 pKa = 4.23HH189 pKa = 6.49FPNGDD194 pKa = 3.68LKK196 pKa = 10.18THH198 pKa = 7.12DD199 pKa = 4.13NLTIFDD205 pKa = 4.08EE206 pKa = 4.73LVAGGADD213 pKa = 3.49VMTTDD218 pKa = 3.77RR219 pKa = 11.84AEE221 pKa = 4.14VLYY224 pKa = 10.24IAHH227 pKa = 7.25EE228 pKa = 4.02YY229 pKa = 11.19DD230 pKa = 3.58EE231 pKa = 5.13LCAVNPDD238 pKa = 3.22EE239 pKa = 5.1TYY241 pKa = 10.95DD242 pKa = 3.93FSVMGYY248 pKa = 7.76MLPQGDD254 pKa = 4.43VVFKK258 pKa = 10.6EE259 pKa = 5.38YY260 pKa = 10.92VDD262 pKa = 3.4QWLTIALKK270 pKa = 10.7DD271 pKa = 3.53GTYY274 pKa = 11.05DD275 pKa = 3.89EE276 pKa = 5.35IAEE279 pKa = 4.11PWVGNVEE286 pKa = 5.61LIPEE290 pKa = 4.37DD291 pKa = 3.36
Molecular weight: 31.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I7MSA4|A0A1I7MSA4_9MICC Adenylosuccinate synthetase OS=Micrococcus terreus OX=574650 GN=purA PE=3 SV=1
MM1 pKa = 7.64PLKK4 pKa = 10.75NLFRR8 pKa = 11.84RR9 pKa = 11.84AQSFARR15 pKa = 11.84SPEE18 pKa = 3.84GKK20 pKa = 10.33RR21 pKa = 11.84MINQAGRR28 pKa = 11.84AARR31 pKa = 11.84SRR33 pKa = 11.84GGRR36 pKa = 11.84RR37 pKa = 11.84GGTRR41 pKa = 11.84PAGGGLMGLAQQFLGRR57 pKa = 11.84GRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 3.22
MM1 pKa = 7.64PLKK4 pKa = 10.75NLFRR8 pKa = 11.84RR9 pKa = 11.84AQSFARR15 pKa = 11.84SPEE18 pKa = 3.84GKK20 pKa = 10.33RR21 pKa = 11.84MINQAGRR28 pKa = 11.84AARR31 pKa = 11.84SRR33 pKa = 11.84GGRR36 pKa = 11.84RR37 pKa = 11.84GGTRR41 pKa = 11.84PAGGGLMGLAQQFLGRR57 pKa = 11.84GRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 3.22
Molecular weight: 6.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
930413 |
41 |
2071 |
337.2 |
36.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.403 ± 0.053 | 0.568 ± 0.01 |
5.925 ± 0.043 | 6.22 ± 0.047 |
2.79 ± 0.03 | 9.045 ± 0.039 |
2.317 ± 0.02 | 3.821 ± 0.036 |
1.994 ± 0.032 | 10.095 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.075 ± 0.022 | 1.907 ± 0.023 |
5.744 ± 0.037 | 3.872 ± 0.028 |
7.158 ± 0.05 | 5.719 ± 0.032 |
6.344 ± 0.038 | 8.599 ± 0.039 |
1.533 ± 0.019 | 1.87 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
