
Chuzan virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus; Palyam virus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
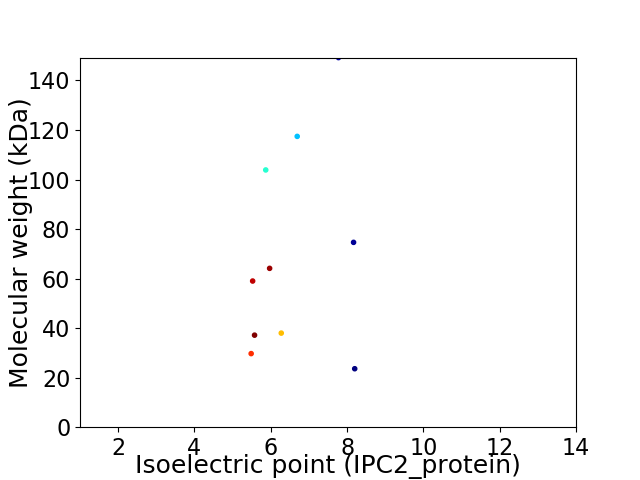
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
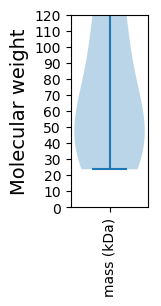
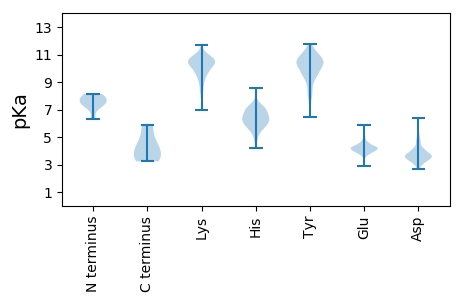
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q9YN53|Q9YN53_9REOV Core protein VP7 OS=Chuzan virus OX=77204 PE=3 SV=1
MM1 pKa = 7.99GDD3 pKa = 3.25RR4 pKa = 11.84KK5 pKa = 10.39RR6 pKa = 11.84YY7 pKa = 6.62TKK9 pKa = 8.78TVCVYY14 pKa = 10.47DD15 pKa = 3.78PQNATFCSRR24 pKa = 11.84GANAGEE30 pKa = 4.3NYY32 pKa = 9.56YY33 pKa = 9.23QVKK36 pKa = 9.83IGRR39 pKa = 11.84TTQITRR45 pKa = 11.84TNAPNPKK52 pKa = 9.53AYY54 pKa = 10.04VVEE57 pKa = 4.19IQRR60 pKa = 11.84PCSVRR65 pKa = 11.84LLDD68 pKa = 4.22GNDD71 pKa = 3.47IISLIISEE79 pKa = 4.45EE80 pKa = 4.42GIEE83 pKa = 4.23VTTEE87 pKa = 3.6RR88 pKa = 11.84WEE90 pKa = 3.97EE91 pKa = 3.82WLFEE95 pKa = 4.3ALTPIPMVISLHH107 pKa = 6.2IKK109 pKa = 8.04GQKK112 pKa = 8.28TEE114 pKa = 4.83AEE116 pKa = 3.88IKK118 pKa = 8.4YY119 pKa = 9.28CKK121 pKa = 10.13ASGVIAPYY129 pKa = 9.47TPNGVDD135 pKa = 3.8RR136 pKa = 11.84RR137 pKa = 11.84VTPQLPGITFTDD149 pKa = 3.5VSVRR153 pKa = 11.84DD154 pKa = 4.16FRR156 pKa = 11.84QKK158 pKa = 9.75MRR160 pKa = 11.84EE161 pKa = 3.78EE162 pKa = 3.94RR163 pKa = 11.84EE164 pKa = 4.03KK165 pKa = 11.01EE166 pKa = 4.04KK167 pKa = 10.85EE168 pKa = 4.07RR169 pKa = 11.84TPMITPIKK177 pKa = 8.79PKK179 pKa = 10.52SEE181 pKa = 4.03APMMGPSTSQQYY193 pKa = 9.77QKK195 pKa = 10.81EE196 pKa = 3.96EE197 pKa = 4.22SIAVGKK203 pKa = 10.64ISDD206 pKa = 3.93YY207 pKa = 10.39VKK209 pKa = 10.76KK210 pKa = 9.63VTLEE214 pKa = 4.23SNVEE218 pKa = 3.89RR219 pKa = 11.84DD220 pKa = 3.28SDD222 pKa = 4.61DD223 pKa = 3.7EE224 pKa = 4.9DD225 pKa = 3.65EE226 pKa = 5.26HH227 pKa = 8.59GSNSDD232 pKa = 3.04SDD234 pKa = 4.28FDD236 pKa = 5.32GEE238 pKa = 4.58PKK240 pKa = 10.67DD241 pKa = 4.01EE242 pKa = 5.34GYY244 pKa = 8.26ITQNYY249 pKa = 7.55MNMIEE254 pKa = 4.27TVKK257 pKa = 10.43MKK259 pKa = 10.74VPGVAGLTLKK269 pKa = 10.35IPKK272 pKa = 9.8AAGKK276 pKa = 7.75TDD278 pKa = 2.97GMIAAKK284 pKa = 8.99KK285 pKa = 9.14CKK287 pKa = 9.11WISVPLFNIDD297 pKa = 4.7KK298 pKa = 10.78DD299 pKa = 3.78GLSYY303 pKa = 11.49DD304 pKa = 4.15FVAVGDD310 pKa = 4.25ANGFLCLRR318 pKa = 11.84DD319 pKa = 3.5GLSYY323 pKa = 11.06LVLPVSGSVHH333 pKa = 5.85
MM1 pKa = 7.99GDD3 pKa = 3.25RR4 pKa = 11.84KK5 pKa = 10.39RR6 pKa = 11.84YY7 pKa = 6.62TKK9 pKa = 8.78TVCVYY14 pKa = 10.47DD15 pKa = 3.78PQNATFCSRR24 pKa = 11.84GANAGEE30 pKa = 4.3NYY32 pKa = 9.56YY33 pKa = 9.23QVKK36 pKa = 9.83IGRR39 pKa = 11.84TTQITRR45 pKa = 11.84TNAPNPKK52 pKa = 9.53AYY54 pKa = 10.04VVEE57 pKa = 4.19IQRR60 pKa = 11.84PCSVRR65 pKa = 11.84LLDD68 pKa = 4.22GNDD71 pKa = 3.47IISLIISEE79 pKa = 4.45EE80 pKa = 4.42GIEE83 pKa = 4.23VTTEE87 pKa = 3.6RR88 pKa = 11.84WEE90 pKa = 3.97EE91 pKa = 3.82WLFEE95 pKa = 4.3ALTPIPMVISLHH107 pKa = 6.2IKK109 pKa = 8.04GQKK112 pKa = 8.28TEE114 pKa = 4.83AEE116 pKa = 3.88IKK118 pKa = 8.4YY119 pKa = 9.28CKK121 pKa = 10.13ASGVIAPYY129 pKa = 9.47TPNGVDD135 pKa = 3.8RR136 pKa = 11.84RR137 pKa = 11.84VTPQLPGITFTDD149 pKa = 3.5VSVRR153 pKa = 11.84DD154 pKa = 4.16FRR156 pKa = 11.84QKK158 pKa = 9.75MRR160 pKa = 11.84EE161 pKa = 3.78EE162 pKa = 3.94RR163 pKa = 11.84EE164 pKa = 4.03KK165 pKa = 11.01EE166 pKa = 4.04KK167 pKa = 10.85EE168 pKa = 4.07RR169 pKa = 11.84TPMITPIKK177 pKa = 8.79PKK179 pKa = 10.52SEE181 pKa = 4.03APMMGPSTSQQYY193 pKa = 9.77QKK195 pKa = 10.81EE196 pKa = 3.96EE197 pKa = 4.22SIAVGKK203 pKa = 10.64ISDD206 pKa = 3.93YY207 pKa = 10.39VKK209 pKa = 10.76KK210 pKa = 9.63VTLEE214 pKa = 4.23SNVEE218 pKa = 3.89RR219 pKa = 11.84DD220 pKa = 3.28SDD222 pKa = 4.61DD223 pKa = 3.7EE224 pKa = 4.9DD225 pKa = 3.65EE226 pKa = 5.26HH227 pKa = 8.59GSNSDD232 pKa = 3.04SDD234 pKa = 4.28FDD236 pKa = 5.32GEE238 pKa = 4.58PKK240 pKa = 10.67DD241 pKa = 4.01EE242 pKa = 5.34GYY244 pKa = 8.26ITQNYY249 pKa = 7.55MNMIEE254 pKa = 4.27TVKK257 pKa = 10.43MKK259 pKa = 10.74VPGVAGLTLKK269 pKa = 10.35IPKK272 pKa = 9.8AAGKK276 pKa = 7.75TDD278 pKa = 2.97GMIAAKK284 pKa = 8.99KK285 pKa = 9.14CKK287 pKa = 9.11WISVPLFNIDD297 pKa = 4.7KK298 pKa = 10.78DD299 pKa = 3.78GLSYY303 pKa = 11.49DD304 pKa = 4.15FVAVGDD310 pKa = 4.25ANGFLCLRR318 pKa = 11.84DD319 pKa = 3.5GLSYY323 pKa = 11.06LVLPVSGSVHH333 pKa = 5.85
Molecular weight: 37.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A109RY65|A0A109RY65_9REOV Minor core protein OS=Chuzan virus OX=77204 PE=4 SV=1
MM1 pKa = 7.71LARR4 pKa = 11.84SLNEE8 pKa = 3.83YY9 pKa = 10.46KK10 pKa = 10.9AMRR13 pKa = 11.84PEE15 pKa = 4.3SEE17 pKa = 4.2MSVVPYY23 pKa = 9.82QPPAYY28 pKa = 6.96PTAPTGTNGEE38 pKa = 4.36KK39 pKa = 10.56DD40 pKa = 3.67FSSISLGVLNNAMNDD55 pKa = 3.34TTAATQAEE63 pKa = 4.48RR64 pKa = 11.84EE65 pKa = 4.28EE66 pKa = 4.31KK67 pKa = 10.1VAYY70 pKa = 10.26ASFAEE75 pKa = 4.11ALRR78 pKa = 11.84DD79 pKa = 3.77SACVRR84 pKa = 11.84EE85 pKa = 4.06IKK87 pKa = 10.45KK88 pKa = 10.18RR89 pKa = 11.84VSSRR93 pKa = 11.84TIVALEE99 pKa = 3.9KK100 pKa = 10.41EE101 pKa = 4.55YY102 pKa = 11.23NHH104 pKa = 6.24QKK106 pKa = 10.38RR107 pKa = 11.84IYY109 pKa = 10.54DD110 pKa = 4.42FIRR113 pKa = 11.84LILFIMSVIAIITSSLSAAIVVIPEE138 pKa = 4.18TKK140 pKa = 10.03ALKK143 pKa = 10.33EE144 pKa = 3.82EE145 pKa = 4.56WISILIHH152 pKa = 5.96VGNLFATGVFVGMSKK167 pKa = 10.23YY168 pKa = 10.7AEE170 pKa = 4.2RR171 pKa = 11.84LDD173 pKa = 4.11SFLKK177 pKa = 9.42RR178 pKa = 11.84TRR180 pKa = 11.84KK181 pKa = 10.09EE182 pKa = 3.7IVKK185 pKa = 9.75KK186 pKa = 10.51RR187 pKa = 11.84SYY189 pKa = 10.75IEE191 pKa = 4.04AANVTWDD198 pKa = 3.81SDD200 pKa = 3.46VDD202 pKa = 3.71TLNFLKK208 pKa = 10.82AAAA211 pKa = 4.4
MM1 pKa = 7.71LARR4 pKa = 11.84SLNEE8 pKa = 3.83YY9 pKa = 10.46KK10 pKa = 10.9AMRR13 pKa = 11.84PEE15 pKa = 4.3SEE17 pKa = 4.2MSVVPYY23 pKa = 9.82QPPAYY28 pKa = 6.96PTAPTGTNGEE38 pKa = 4.36KK39 pKa = 10.56DD40 pKa = 3.67FSSISLGVLNNAMNDD55 pKa = 3.34TTAATQAEE63 pKa = 4.48RR64 pKa = 11.84EE65 pKa = 4.28EE66 pKa = 4.31KK67 pKa = 10.1VAYY70 pKa = 10.26ASFAEE75 pKa = 4.11ALRR78 pKa = 11.84DD79 pKa = 3.77SACVRR84 pKa = 11.84EE85 pKa = 4.06IKK87 pKa = 10.45KK88 pKa = 10.18RR89 pKa = 11.84VSSRR93 pKa = 11.84TIVALEE99 pKa = 3.9KK100 pKa = 10.41EE101 pKa = 4.55YY102 pKa = 11.23NHH104 pKa = 6.24QKK106 pKa = 10.38RR107 pKa = 11.84IYY109 pKa = 10.54DD110 pKa = 4.42FIRR113 pKa = 11.84LILFIMSVIAIITSSLSAAIVVIPEE138 pKa = 4.18TKK140 pKa = 10.03ALKK143 pKa = 10.33EE144 pKa = 3.82EE145 pKa = 4.56WISILIHH152 pKa = 5.96VGNLFATGVFVGMSKK167 pKa = 10.23YY168 pKa = 10.7AEE170 pKa = 4.2RR171 pKa = 11.84LDD173 pKa = 4.11SFLKK177 pKa = 9.42RR178 pKa = 11.84TRR180 pKa = 11.84KK181 pKa = 10.09EE182 pKa = 3.7IVKK185 pKa = 9.75KK186 pKa = 10.51RR187 pKa = 11.84SYY189 pKa = 10.75IEE191 pKa = 4.04AANVTWDD198 pKa = 3.81SDD200 pKa = 3.46VDD202 pKa = 3.71TLNFLKK208 pKa = 10.82AAAA211 pKa = 4.4
Molecular weight: 23.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6071 |
211 |
1295 |
607.1 |
69.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.111 ± 0.632 | 1.219 ± 0.214 |
6.078 ± 0.258 | 6.968 ± 0.445 |
4.414 ± 0.368 | 5.732 ± 0.483 |
2.026 ± 0.207 | 7.478 ± 0.25 |
5.93 ± 0.646 | 8.285 ± 0.435 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.624 ± 0.301 | 4.167 ± 0.26 |
3.838 ± 0.293 | 3.821 ± 0.311 |
6.951 ± 0.336 | 6.473 ± 0.299 |
5.222 ± 0.396 | 6.342 ± 0.217 |
1.268 ± 0.236 | 4.052 ± 0.31 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
