
Simian foamy virus Pongo pygmaeus pygmaeus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Spumaretrovirinae; Simiispumavirus; Bornean orangutan simian foamy virus
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
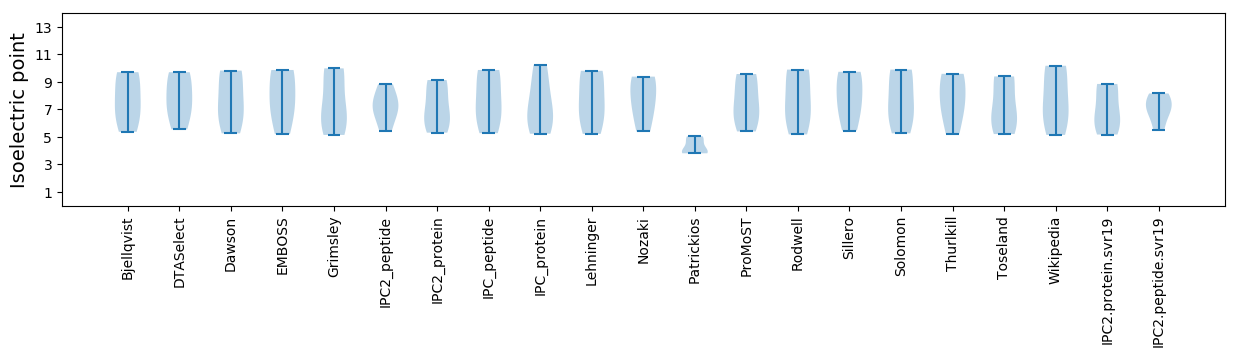
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q7SIS5|Q7SIS5_9RETR Env leader protein OS=Simian foamy virus Pongo pygmaeus pygmaeus OX=221703 GN=env PE=4 SV=1
MM1 pKa = 8.13DD2 pKa = 5.82PNQEE6 pKa = 4.07EE7 pKa = 4.61EE8 pKa = 4.48PVAGTSGMNQDD19 pKa = 4.06PVPFIPEE26 pKa = 4.72GIAAANEE33 pKa = 4.18SDD35 pKa = 4.84DD36 pKa = 4.31EE37 pKa = 4.3EE38 pKa = 5.71PEE40 pKa = 3.89QFLYY44 pKa = 10.37KK45 pKa = 10.36VYY47 pKa = 10.62QEE49 pKa = 4.31SVKK52 pKa = 10.86KK53 pKa = 10.8NGGDD57 pKa = 3.68YY58 pKa = 10.54PKK60 pKa = 10.86LEE62 pKa = 4.25DD63 pKa = 4.39WIPSPEE69 pKa = 3.99EE70 pKa = 3.62MSKK73 pKa = 10.46SVCISLILTCLYY85 pKa = 10.26NAEE88 pKa = 4.2KK89 pKa = 10.37AAQIKK94 pKa = 8.87DD95 pKa = 2.84WGYY98 pKa = 9.42IVHH101 pKa = 6.92WEE103 pKa = 3.96QSPTDD108 pKa = 3.57SKK110 pKa = 11.74YY111 pKa = 10.73FLIKK115 pKa = 10.02YY116 pKa = 6.86EE117 pKa = 4.74CPMCDD122 pKa = 4.54SINQEE127 pKa = 4.56PMPIWWDD134 pKa = 3.39DD135 pKa = 3.4RR136 pKa = 11.84LKK138 pKa = 10.64LWRR141 pKa = 11.84KK142 pKa = 6.36MGCRR146 pKa = 11.84AVMGSIVYY154 pKa = 9.91ALKK157 pKa = 10.91NHH159 pKa = 5.68VDD161 pKa = 3.17KK162 pKa = 11.38CNSQVHH168 pKa = 5.96PLRR171 pKa = 11.84KK172 pKa = 8.01TGNRR176 pKa = 11.84RR177 pKa = 11.84PRR179 pKa = 11.84PRR181 pKa = 11.84IDD183 pKa = 3.95PIRR186 pKa = 11.84RR187 pKa = 11.84CNRR190 pKa = 11.84LTGNYY195 pKa = 8.92VPGRR199 pKa = 11.84RR200 pKa = 11.84GSTKK204 pKa = 9.48PSNPSSHH211 pKa = 7.06PSSGIPLAPGPRR223 pKa = 11.84QCSTNTSNPPEE234 pKa = 4.17SLLRR238 pKa = 11.84LLPGNDD244 pKa = 4.0AISPALAISMSGGQIWEE261 pKa = 4.04EE262 pKa = 4.5VYY264 pKa = 11.4NDD266 pKa = 4.21LLLDD270 pKa = 3.93ATLGTSDD277 pKa = 3.25NN278 pKa = 4.01
MM1 pKa = 8.13DD2 pKa = 5.82PNQEE6 pKa = 4.07EE7 pKa = 4.61EE8 pKa = 4.48PVAGTSGMNQDD19 pKa = 4.06PVPFIPEE26 pKa = 4.72GIAAANEE33 pKa = 4.18SDD35 pKa = 4.84DD36 pKa = 4.31EE37 pKa = 4.3EE38 pKa = 5.71PEE40 pKa = 3.89QFLYY44 pKa = 10.37KK45 pKa = 10.36VYY47 pKa = 10.62QEE49 pKa = 4.31SVKK52 pKa = 10.86KK53 pKa = 10.8NGGDD57 pKa = 3.68YY58 pKa = 10.54PKK60 pKa = 10.86LEE62 pKa = 4.25DD63 pKa = 4.39WIPSPEE69 pKa = 3.99EE70 pKa = 3.62MSKK73 pKa = 10.46SVCISLILTCLYY85 pKa = 10.26NAEE88 pKa = 4.2KK89 pKa = 10.37AAQIKK94 pKa = 8.87DD95 pKa = 2.84WGYY98 pKa = 9.42IVHH101 pKa = 6.92WEE103 pKa = 3.96QSPTDD108 pKa = 3.57SKK110 pKa = 11.74YY111 pKa = 10.73FLIKK115 pKa = 10.02YY116 pKa = 6.86EE117 pKa = 4.74CPMCDD122 pKa = 4.54SINQEE127 pKa = 4.56PMPIWWDD134 pKa = 3.39DD135 pKa = 3.4RR136 pKa = 11.84LKK138 pKa = 10.64LWRR141 pKa = 11.84KK142 pKa = 6.36MGCRR146 pKa = 11.84AVMGSIVYY154 pKa = 9.91ALKK157 pKa = 10.91NHH159 pKa = 5.68VDD161 pKa = 3.17KK162 pKa = 11.38CNSQVHH168 pKa = 5.96PLRR171 pKa = 11.84KK172 pKa = 8.01TGNRR176 pKa = 11.84RR177 pKa = 11.84PRR179 pKa = 11.84PRR181 pKa = 11.84IDD183 pKa = 3.95PIRR186 pKa = 11.84RR187 pKa = 11.84CNRR190 pKa = 11.84LTGNYY195 pKa = 8.92VPGRR199 pKa = 11.84RR200 pKa = 11.84GSTKK204 pKa = 9.48PSNPSSHH211 pKa = 7.06PSSGIPLAPGPRR223 pKa = 11.84QCSTNTSNPPEE234 pKa = 4.17SLLRR238 pKa = 11.84LLPGNDD244 pKa = 4.0AISPALAISMSGGQIWEE261 pKa = 4.04EE262 pKa = 4.5VYY264 pKa = 11.4NDD266 pKa = 4.21LLLDD270 pKa = 3.93ATLGTSDD277 pKa = 3.25NN278 pKa = 4.01
Molecular weight: 31.13 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q7SIS7|Q7SIS7_9RETR Gag polyprotein OS=Simian foamy virus Pongo pygmaeus pygmaeus OX=221703 GN=gag PE=4 SV=1
MM1 pKa = 7.61AAQNFEE7 pKa = 4.86LDD9 pKa = 3.77VQEE12 pKa = 5.2LLNLFQDD19 pKa = 3.68NGVTRR24 pKa = 11.84NPRR27 pKa = 11.84HH28 pKa = 6.44LEE30 pKa = 4.23TIGLRR35 pKa = 11.84MLGGWWGEE43 pKa = 3.78QEE45 pKa = 4.58RR46 pKa = 11.84YY47 pKa = 8.37QSARR51 pKa = 11.84IILQDD56 pKa = 4.13DD57 pKa = 3.93DD58 pKa = 5.43GEE60 pKa = 4.28PLQVPRR66 pKa = 11.84WEE68 pKa = 4.18EE69 pKa = 3.35VLRR72 pKa = 11.84PVNPLAHH79 pKa = 6.14FVISAPWDD87 pKa = 3.31QLRR90 pKa = 11.84RR91 pKa = 11.84AFHH94 pKa = 6.97DD95 pKa = 3.82LDD97 pKa = 3.8VGNGALRR104 pKa = 11.84FGPLANGNYY113 pKa = 9.41IPEE116 pKa = 4.63DD117 pKa = 4.24PYY119 pKa = 9.74STSYY123 pKa = 11.02RR124 pKa = 11.84PVNPQEE130 pKa = 3.91MAQMQRR136 pKa = 11.84DD137 pKa = 3.7EE138 pKa = 4.6LEE140 pKa = 4.14EE141 pKa = 3.84VLEE144 pKa = 4.17VQGEE148 pKa = 4.35IEE150 pKa = 4.38LQMIDD155 pKa = 4.71LIEE158 pKa = 4.2MQTIEE163 pKa = 4.6IRR165 pKa = 11.84GLRR168 pKa = 11.84QLVNEE173 pKa = 4.6LQRR176 pKa = 11.84EE177 pKa = 4.02RR178 pKa = 11.84DD179 pKa = 3.49SGRR182 pKa = 11.84GASIPGASSSPPPQSYY198 pKa = 10.69VGLPGMPSVSGLSSLQPEE216 pKa = 4.57ASSTPGGRR224 pKa = 11.84APRR227 pKa = 11.84VSFHH231 pKa = 7.03PSNPFVRR238 pKa = 11.84PPSPEE243 pKa = 3.61RR244 pKa = 11.84PRR246 pKa = 11.84ARR248 pKa = 11.84SSEE251 pKa = 4.42RR252 pKa = 11.84IPLPLQPPVIQYY264 pKa = 9.9VPVPQPVLAPAPAPSPVIPIQHH286 pKa = 5.76IRR288 pKa = 11.84AVTGEE293 pKa = 4.25VPNNPRR299 pKa = 11.84DD300 pKa = 3.38IPMWIGRR307 pKa = 11.84NAPAIEE313 pKa = 4.1GVYY316 pKa = 10.33PVTTPDD322 pKa = 2.9LRR324 pKa = 11.84ARR326 pKa = 11.84IINALIGGKK335 pKa = 9.78SGIHH339 pKa = 4.87LTAPEE344 pKa = 4.03AVTWASAVAAIFTRR358 pKa = 11.84THH360 pKa = 6.36GSFPMHH366 pKa = 6.49NLSAILTGIANGEE379 pKa = 4.37GVEE382 pKa = 4.23SAYY385 pKa = 11.12NLGMMLSNGDD395 pKa = 3.52FNLVYY400 pKa = 10.51GIVRR404 pKa = 11.84GLLPGQAAVAYY415 pKa = 6.92MQQRR419 pKa = 11.84LDD421 pKa = 4.24AEE423 pKa = 4.42PSDD426 pKa = 4.09ALRR429 pKa = 11.84AQNFIQHH436 pKa = 6.11LHH438 pKa = 5.71LVYY441 pKa = 10.58EE442 pKa = 4.33ILGLNHH448 pKa = 7.38RR449 pKa = 11.84GQSIRR454 pKa = 11.84TSLPTSTRR462 pKa = 11.84PRR464 pKa = 11.84GQGRR468 pKa = 11.84GRR470 pKa = 11.84GQGQGSIPSTPRR482 pKa = 11.84RR483 pKa = 11.84PQSGRR488 pKa = 11.84GLSTPNRR495 pKa = 11.84GSNNANNNTQSNVQTEE511 pKa = 4.43TPRR514 pKa = 11.84RR515 pKa = 11.84SFGGYY520 pKa = 7.35NLRR523 pKa = 11.84PNTFRR528 pKa = 11.84PQRR531 pKa = 11.84YY532 pKa = 9.31GGGQGQRR539 pKa = 11.84RR540 pKa = 11.84DD541 pKa = 3.68SQPDD545 pKa = 3.03RR546 pKa = 11.84RR547 pKa = 11.84SQGSSQNNRR556 pKa = 11.84PSAPLEE562 pKa = 4.04SRR564 pKa = 11.84GEE566 pKa = 3.96QSRR569 pKa = 11.84GPGGGGRR576 pKa = 11.84AGGRR580 pKa = 11.84RR581 pKa = 11.84NQNRR585 pKa = 11.84NSGQGNEE592 pKa = 4.35SSSHH596 pKa = 5.48AVNAVTQSAVTEE608 pKa = 4.12QQNEE612 pKa = 4.63SPTPPPTSGGRR623 pKa = 11.84SS624 pKa = 3.24
MM1 pKa = 7.61AAQNFEE7 pKa = 4.86LDD9 pKa = 3.77VQEE12 pKa = 5.2LLNLFQDD19 pKa = 3.68NGVTRR24 pKa = 11.84NPRR27 pKa = 11.84HH28 pKa = 6.44LEE30 pKa = 4.23TIGLRR35 pKa = 11.84MLGGWWGEE43 pKa = 3.78QEE45 pKa = 4.58RR46 pKa = 11.84YY47 pKa = 8.37QSARR51 pKa = 11.84IILQDD56 pKa = 4.13DD57 pKa = 3.93DD58 pKa = 5.43GEE60 pKa = 4.28PLQVPRR66 pKa = 11.84WEE68 pKa = 4.18EE69 pKa = 3.35VLRR72 pKa = 11.84PVNPLAHH79 pKa = 6.14FVISAPWDD87 pKa = 3.31QLRR90 pKa = 11.84RR91 pKa = 11.84AFHH94 pKa = 6.97DD95 pKa = 3.82LDD97 pKa = 3.8VGNGALRR104 pKa = 11.84FGPLANGNYY113 pKa = 9.41IPEE116 pKa = 4.63DD117 pKa = 4.24PYY119 pKa = 9.74STSYY123 pKa = 11.02RR124 pKa = 11.84PVNPQEE130 pKa = 3.91MAQMQRR136 pKa = 11.84DD137 pKa = 3.7EE138 pKa = 4.6LEE140 pKa = 4.14EE141 pKa = 3.84VLEE144 pKa = 4.17VQGEE148 pKa = 4.35IEE150 pKa = 4.38LQMIDD155 pKa = 4.71LIEE158 pKa = 4.2MQTIEE163 pKa = 4.6IRR165 pKa = 11.84GLRR168 pKa = 11.84QLVNEE173 pKa = 4.6LQRR176 pKa = 11.84EE177 pKa = 4.02RR178 pKa = 11.84DD179 pKa = 3.49SGRR182 pKa = 11.84GASIPGASSSPPPQSYY198 pKa = 10.69VGLPGMPSVSGLSSLQPEE216 pKa = 4.57ASSTPGGRR224 pKa = 11.84APRR227 pKa = 11.84VSFHH231 pKa = 7.03PSNPFVRR238 pKa = 11.84PPSPEE243 pKa = 3.61RR244 pKa = 11.84PRR246 pKa = 11.84ARR248 pKa = 11.84SSEE251 pKa = 4.42RR252 pKa = 11.84IPLPLQPPVIQYY264 pKa = 9.9VPVPQPVLAPAPAPSPVIPIQHH286 pKa = 5.76IRR288 pKa = 11.84AVTGEE293 pKa = 4.25VPNNPRR299 pKa = 11.84DD300 pKa = 3.38IPMWIGRR307 pKa = 11.84NAPAIEE313 pKa = 4.1GVYY316 pKa = 10.33PVTTPDD322 pKa = 2.9LRR324 pKa = 11.84ARR326 pKa = 11.84IINALIGGKK335 pKa = 9.78SGIHH339 pKa = 4.87LTAPEE344 pKa = 4.03AVTWASAVAAIFTRR358 pKa = 11.84THH360 pKa = 6.36GSFPMHH366 pKa = 6.49NLSAILTGIANGEE379 pKa = 4.37GVEE382 pKa = 4.23SAYY385 pKa = 11.12NLGMMLSNGDD395 pKa = 3.52FNLVYY400 pKa = 10.51GIVRR404 pKa = 11.84GLLPGQAAVAYY415 pKa = 6.92MQQRR419 pKa = 11.84LDD421 pKa = 4.24AEE423 pKa = 4.42PSDD426 pKa = 4.09ALRR429 pKa = 11.84AQNFIQHH436 pKa = 6.11LHH438 pKa = 5.71LVYY441 pKa = 10.58EE442 pKa = 4.33ILGLNHH448 pKa = 7.38RR449 pKa = 11.84GQSIRR454 pKa = 11.84TSLPTSTRR462 pKa = 11.84PRR464 pKa = 11.84GQGRR468 pKa = 11.84GRR470 pKa = 11.84GQGQGSIPSTPRR482 pKa = 11.84RR483 pKa = 11.84PQSGRR488 pKa = 11.84GLSTPNRR495 pKa = 11.84GSNNANNNTQSNVQTEE511 pKa = 4.43TPRR514 pKa = 11.84RR515 pKa = 11.84SFGGYY520 pKa = 7.35NLRR523 pKa = 11.84PNTFRR528 pKa = 11.84PQRR531 pKa = 11.84YY532 pKa = 9.31GGGQGQRR539 pKa = 11.84RR540 pKa = 11.84DD541 pKa = 3.68SQPDD545 pKa = 3.03RR546 pKa = 11.84RR547 pKa = 11.84SQGSSQNNRR556 pKa = 11.84PSAPLEE562 pKa = 4.04SRR564 pKa = 11.84GEE566 pKa = 3.96QSRR569 pKa = 11.84GPGGGGRR576 pKa = 11.84AGGRR580 pKa = 11.84RR581 pKa = 11.84NQNRR585 pKa = 11.84NSGQGNEE592 pKa = 4.35SSSHH596 pKa = 5.48AVNAVTQSAVTEE608 pKa = 4.12QQNEE612 pKa = 4.63SPTPPPTSGGRR623 pKa = 11.84SS624 pKa = 3.24
Molecular weight: 67.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3357 |
278 |
1145 |
671.4 |
75.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.541 ± 0.631 | 1.549 ± 0.64 |
4.349 ± 0.448 | 5.749 ± 0.505 |
2.532 ± 0.198 | 6.136 ± 1.284 |
2.562 ± 0.322 | 6.673 ± 0.858 |
5.809 ± 1.34 | 9.89 ± 0.679 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.906 ± 0.286 | 5.451 ± 0.249 |
7.417 ± 0.869 | 5.451 ± 0.489 |
5.004 ± 1.158 | 6.494 ± 0.611 |
5.898 ± 0.62 | 5.839 ± 0.48 |
1.906 ± 0.241 | 3.843 ± 0.509 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
