
Saccharomonospora phage PIS 136
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
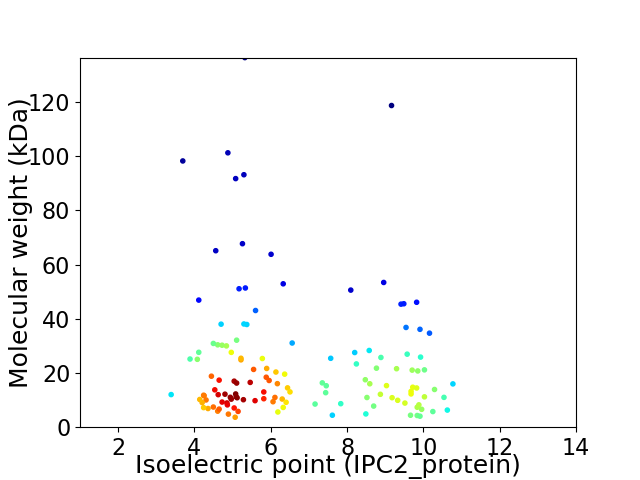
Virtual 2D-PAGE plot for 132 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I4AZD6|I4AZD6_9CAUD Uncharacterized protein OS=Saccharomonospora phage PIS 136 OX=182851 GN=PIS_003 PE=4 SV=1
MM1 pKa = 7.8SYY3 pKa = 10.28QGNTGCSCGGGGTSSGGGSGGGTTVVCSRR32 pKa = 11.84PSIPVAVPGMCLSDD46 pKa = 3.48GTPIATISTRR56 pKa = 11.84AKK58 pKa = 9.88DD59 pKa = 3.89GTWVRR64 pKa = 11.84TGWLNLSTGGFTAGAPPASAKK85 pKa = 9.98PCNPSDD91 pKa = 3.65NGGGEE96 pKa = 4.22PATLAGVHH104 pKa = 5.57VEE106 pKa = 4.31DD107 pKa = 4.3WCDD110 pKa = 3.25VDD112 pKa = 5.88DD113 pKa = 5.68DD114 pKa = 5.41GNVLSPVLAKK124 pKa = 11.01YY125 pKa = 7.62EE126 pKa = 4.08TDD128 pKa = 3.51DD129 pKa = 3.25NGAIVGVEE137 pKa = 3.83FLTPDD142 pKa = 3.76GQPYY146 pKa = 9.68EE147 pKa = 4.06IQGTLGICNTDD158 pKa = 3.04GPADD162 pKa = 3.56LTGRR166 pKa = 11.84EE167 pKa = 4.32LVRR170 pKa = 11.84LCDD173 pKa = 3.6VADD176 pKa = 4.97DD177 pKa = 3.89GTITVFVRR185 pKa = 11.84DD186 pKa = 3.97YY187 pKa = 11.09EE188 pKa = 4.28RR189 pKa = 11.84DD190 pKa = 3.36SAGAVVGFTDD200 pKa = 4.52YY201 pKa = 11.28DD202 pKa = 3.44IDD204 pKa = 4.05GQPYY208 pKa = 8.13TPSGEE213 pKa = 4.28VTSCDD218 pKa = 3.07AAQTEE223 pKa = 4.82EE224 pKa = 4.53EE225 pKa = 4.78CASPTTPLATTGLCLADD242 pKa = 3.69GTPIATVSTRR252 pKa = 11.84DD253 pKa = 3.24CDD255 pKa = 4.07GVVSTDD261 pKa = 2.51GWINLLTNTYY271 pKa = 8.76TAGQPPAGARR281 pKa = 11.84ACGEE285 pKa = 3.79TSAFDD290 pKa = 3.67VSGVLCDD297 pKa = 3.73VNEE300 pKa = 4.51SGDD303 pKa = 3.65VLGLVLIEE311 pKa = 4.17VEE313 pKa = 4.32RR314 pKa = 11.84GTDD317 pKa = 3.74GEE319 pKa = 4.37ITGTRR324 pKa = 11.84LINAADD330 pKa = 3.68GSEE333 pKa = 4.03YY334 pKa = 10.48TLQGEE339 pKa = 4.58LTVCPAGVDD348 pKa = 3.44QPEE351 pKa = 3.79RR352 pKa = 11.84DD353 pKa = 4.09LIEE356 pKa = 5.05LCDD359 pKa = 3.9LTDD362 pKa = 4.42DD363 pKa = 5.15GEE365 pKa = 4.76GTTATRR371 pKa = 11.84FLRR374 pKa = 11.84DD375 pKa = 3.16YY376 pKa = 11.53ARR378 pKa = 11.84DD379 pKa = 3.59EE380 pKa = 4.25NGTIVGYY387 pKa = 11.08ADD389 pKa = 3.51YY390 pKa = 9.69TLSGDD395 pKa = 3.78PYY397 pKa = 9.13TPSGNVGSCAAGAAQDD413 pKa = 4.13PQDD416 pKa = 3.59HH417 pKa = 6.56YY418 pKa = 11.43LVQGHH423 pKa = 6.31PMCVVDD429 pKa = 5.23ADD431 pKa = 3.74NAIVRR436 pKa = 11.84RR437 pKa = 11.84VLSEE441 pKa = 3.89HH442 pKa = 6.83VYY444 pKa = 10.86DD445 pKa = 3.58ATTGEE450 pKa = 4.24RR451 pKa = 11.84VEE453 pKa = 4.18TRR455 pKa = 11.84IVDD458 pKa = 3.78PVTGEE463 pKa = 4.04PVDD466 pKa = 4.31LGAGEE471 pKa = 4.45TVADD475 pKa = 4.39CADD478 pKa = 3.42AAADD482 pKa = 5.31DD483 pKa = 4.48EE484 pKa = 4.77PCRR487 pKa = 11.84NTATLMLCDD496 pKa = 4.16VVTDD500 pKa = 4.76DD501 pKa = 5.69SDD503 pKa = 4.61DD504 pKa = 4.48PEE506 pKa = 4.46PTWVQVDD513 pKa = 4.46FEE515 pKa = 4.54QLPTTPPAYY524 pKa = 9.52TGSLPNGVTFTVQGGMPAPRR544 pKa = 11.84GYY546 pKa = 10.43RR547 pKa = 11.84FSNAHH552 pKa = 5.95SNAPDD557 pKa = 4.14GPQRR561 pKa = 11.84WNFDD565 pKa = 3.22QPVMVRR571 pKa = 11.84VGITDD576 pKa = 3.36VQTQLTTNPNGWHH589 pKa = 5.4EE590 pKa = 4.39CVRR593 pKa = 11.84VDD595 pKa = 5.29SKK597 pKa = 11.68LEE599 pKa = 3.83PEE601 pKa = 4.96GVHH604 pKa = 6.9SLHH607 pKa = 6.68EE608 pKa = 3.96WDD610 pKa = 3.32ATNRR614 pKa = 11.84TLCGTADD621 pKa = 3.57TEE623 pKa = 4.65GGGGEE628 pKa = 4.22AVTVFASSEE637 pKa = 4.27PITEE641 pKa = 4.22FEE643 pKa = 5.92LIDD646 pKa = 4.37LTTNPNNQVVGVSTSFVEE664 pKa = 4.37VLLPASGDD672 pKa = 3.72EE673 pKa = 4.25PNVTVTRR680 pKa = 11.84FLRR683 pKa = 11.84HH684 pKa = 5.9IVTDD688 pKa = 4.02CATGEE693 pKa = 4.06IVSINDD699 pKa = 3.27TTLDD703 pKa = 3.73GEE705 pKa = 4.48PYY707 pKa = 9.71EE708 pKa = 4.32VTGEE712 pKa = 4.15VEE714 pKa = 3.98QCQIEE719 pKa = 4.61CGSSCNITPGAPGAPGSPTDD739 pKa = 4.08PQPCQNVTTILLCDD753 pKa = 4.38TIDD756 pKa = 4.89DD757 pKa = 4.23GDD759 pKa = 4.25GEE761 pKa = 4.48QVVPFLRR768 pKa = 11.84SYY770 pKa = 11.1QIDD773 pKa = 3.87NDD775 pKa = 3.76SGTITGHH782 pKa = 6.43TDD784 pKa = 3.1TEE786 pKa = 4.93LDD788 pKa = 3.04GSTPYY793 pKa = 10.49EE794 pKa = 3.92PQGEE798 pKa = 4.41VGQCQPASSGADD810 pKa = 3.42PLPEE814 pKa = 4.64FDD816 pKa = 3.6VAPIVLCDD824 pKa = 3.59TVDD827 pKa = 3.46GGEE830 pKa = 4.32GEE832 pKa = 4.23QAVEE836 pKa = 3.79FLRR839 pKa = 11.84FFRR842 pKa = 11.84VDD844 pKa = 3.01NSTGEE849 pKa = 4.46VTGHH853 pKa = 6.67TDD855 pKa = 3.25TEE857 pKa = 4.66LDD859 pKa = 4.05GITPYY864 pKa = 10.65EE865 pKa = 3.98PQGEE869 pKa = 4.39VGQCQSGGEE878 pKa = 4.25CCEE881 pKa = 4.1PPEE884 pKa = 4.59PVPSLDD890 pKa = 3.23IRR892 pKa = 11.84CEE894 pKa = 4.1TVVLCDD900 pKa = 3.88TGPDD904 pKa = 3.48GDD906 pKa = 4.85DD907 pKa = 3.17PVSFMRR913 pKa = 11.84AICRR917 pKa = 11.84DD918 pKa = 2.88SSGNVVSVTDD928 pKa = 3.76TEE930 pKa = 4.53LDD932 pKa = 3.15AVTPYY937 pKa = 9.9TVRR940 pKa = 11.84SS941 pKa = 3.62
MM1 pKa = 7.8SYY3 pKa = 10.28QGNTGCSCGGGGTSSGGGSGGGTTVVCSRR32 pKa = 11.84PSIPVAVPGMCLSDD46 pKa = 3.48GTPIATISTRR56 pKa = 11.84AKK58 pKa = 9.88DD59 pKa = 3.89GTWVRR64 pKa = 11.84TGWLNLSTGGFTAGAPPASAKK85 pKa = 9.98PCNPSDD91 pKa = 3.65NGGGEE96 pKa = 4.22PATLAGVHH104 pKa = 5.57VEE106 pKa = 4.31DD107 pKa = 4.3WCDD110 pKa = 3.25VDD112 pKa = 5.88DD113 pKa = 5.68DD114 pKa = 5.41GNVLSPVLAKK124 pKa = 11.01YY125 pKa = 7.62EE126 pKa = 4.08TDD128 pKa = 3.51DD129 pKa = 3.25NGAIVGVEE137 pKa = 3.83FLTPDD142 pKa = 3.76GQPYY146 pKa = 9.68EE147 pKa = 4.06IQGTLGICNTDD158 pKa = 3.04GPADD162 pKa = 3.56LTGRR166 pKa = 11.84EE167 pKa = 4.32LVRR170 pKa = 11.84LCDD173 pKa = 3.6VADD176 pKa = 4.97DD177 pKa = 3.89GTITVFVRR185 pKa = 11.84DD186 pKa = 3.97YY187 pKa = 11.09EE188 pKa = 4.28RR189 pKa = 11.84DD190 pKa = 3.36SAGAVVGFTDD200 pKa = 4.52YY201 pKa = 11.28DD202 pKa = 3.44IDD204 pKa = 4.05GQPYY208 pKa = 8.13TPSGEE213 pKa = 4.28VTSCDD218 pKa = 3.07AAQTEE223 pKa = 4.82EE224 pKa = 4.53EE225 pKa = 4.78CASPTTPLATTGLCLADD242 pKa = 3.69GTPIATVSTRR252 pKa = 11.84DD253 pKa = 3.24CDD255 pKa = 4.07GVVSTDD261 pKa = 2.51GWINLLTNTYY271 pKa = 8.76TAGQPPAGARR281 pKa = 11.84ACGEE285 pKa = 3.79TSAFDD290 pKa = 3.67VSGVLCDD297 pKa = 3.73VNEE300 pKa = 4.51SGDD303 pKa = 3.65VLGLVLIEE311 pKa = 4.17VEE313 pKa = 4.32RR314 pKa = 11.84GTDD317 pKa = 3.74GEE319 pKa = 4.37ITGTRR324 pKa = 11.84LINAADD330 pKa = 3.68GSEE333 pKa = 4.03YY334 pKa = 10.48TLQGEE339 pKa = 4.58LTVCPAGVDD348 pKa = 3.44QPEE351 pKa = 3.79RR352 pKa = 11.84DD353 pKa = 4.09LIEE356 pKa = 5.05LCDD359 pKa = 3.9LTDD362 pKa = 4.42DD363 pKa = 5.15GEE365 pKa = 4.76GTTATRR371 pKa = 11.84FLRR374 pKa = 11.84DD375 pKa = 3.16YY376 pKa = 11.53ARR378 pKa = 11.84DD379 pKa = 3.59EE380 pKa = 4.25NGTIVGYY387 pKa = 11.08ADD389 pKa = 3.51YY390 pKa = 9.69TLSGDD395 pKa = 3.78PYY397 pKa = 9.13TPSGNVGSCAAGAAQDD413 pKa = 4.13PQDD416 pKa = 3.59HH417 pKa = 6.56YY418 pKa = 11.43LVQGHH423 pKa = 6.31PMCVVDD429 pKa = 5.23ADD431 pKa = 3.74NAIVRR436 pKa = 11.84RR437 pKa = 11.84VLSEE441 pKa = 3.89HH442 pKa = 6.83VYY444 pKa = 10.86DD445 pKa = 3.58ATTGEE450 pKa = 4.24RR451 pKa = 11.84VEE453 pKa = 4.18TRR455 pKa = 11.84IVDD458 pKa = 3.78PVTGEE463 pKa = 4.04PVDD466 pKa = 4.31LGAGEE471 pKa = 4.45TVADD475 pKa = 4.39CADD478 pKa = 3.42AAADD482 pKa = 5.31DD483 pKa = 4.48EE484 pKa = 4.77PCRR487 pKa = 11.84NTATLMLCDD496 pKa = 4.16VVTDD500 pKa = 4.76DD501 pKa = 5.69SDD503 pKa = 4.61DD504 pKa = 4.48PEE506 pKa = 4.46PTWVQVDD513 pKa = 4.46FEE515 pKa = 4.54QLPTTPPAYY524 pKa = 9.52TGSLPNGVTFTVQGGMPAPRR544 pKa = 11.84GYY546 pKa = 10.43RR547 pKa = 11.84FSNAHH552 pKa = 5.95SNAPDD557 pKa = 4.14GPQRR561 pKa = 11.84WNFDD565 pKa = 3.22QPVMVRR571 pKa = 11.84VGITDD576 pKa = 3.36VQTQLTTNPNGWHH589 pKa = 5.4EE590 pKa = 4.39CVRR593 pKa = 11.84VDD595 pKa = 5.29SKK597 pKa = 11.68LEE599 pKa = 3.83PEE601 pKa = 4.96GVHH604 pKa = 6.9SLHH607 pKa = 6.68EE608 pKa = 3.96WDD610 pKa = 3.32ATNRR614 pKa = 11.84TLCGTADD621 pKa = 3.57TEE623 pKa = 4.65GGGGEE628 pKa = 4.22AVTVFASSEE637 pKa = 4.27PITEE641 pKa = 4.22FEE643 pKa = 5.92LIDD646 pKa = 4.37LTTNPNNQVVGVSTSFVEE664 pKa = 4.37VLLPASGDD672 pKa = 3.72EE673 pKa = 4.25PNVTVTRR680 pKa = 11.84FLRR683 pKa = 11.84HH684 pKa = 5.9IVTDD688 pKa = 4.02CATGEE693 pKa = 4.06IVSINDD699 pKa = 3.27TTLDD703 pKa = 3.73GEE705 pKa = 4.48PYY707 pKa = 9.71EE708 pKa = 4.32VTGEE712 pKa = 4.15VEE714 pKa = 3.98QCQIEE719 pKa = 4.61CGSSCNITPGAPGAPGSPTDD739 pKa = 4.08PQPCQNVTTILLCDD753 pKa = 4.38TIDD756 pKa = 4.89DD757 pKa = 4.23GDD759 pKa = 4.25GEE761 pKa = 4.48QVVPFLRR768 pKa = 11.84SYY770 pKa = 11.1QIDD773 pKa = 3.87NDD775 pKa = 3.76SGTITGHH782 pKa = 6.43TDD784 pKa = 3.1TEE786 pKa = 4.93LDD788 pKa = 3.04GSTPYY793 pKa = 10.49EE794 pKa = 3.92PQGEE798 pKa = 4.41VGQCQPASSGADD810 pKa = 3.42PLPEE814 pKa = 4.64FDD816 pKa = 3.6VAPIVLCDD824 pKa = 3.59TVDD827 pKa = 3.46GGEE830 pKa = 4.32GEE832 pKa = 4.23QAVEE836 pKa = 3.79FLRR839 pKa = 11.84FFRR842 pKa = 11.84VDD844 pKa = 3.01NSTGEE849 pKa = 4.46VTGHH853 pKa = 6.67TDD855 pKa = 3.25TEE857 pKa = 4.66LDD859 pKa = 4.05GITPYY864 pKa = 10.65EE865 pKa = 3.98PQGEE869 pKa = 4.39VGQCQSGGEE878 pKa = 4.25CCEE881 pKa = 4.1PPEE884 pKa = 4.59PVPSLDD890 pKa = 3.23IRR892 pKa = 11.84CEE894 pKa = 4.1TVVLCDD900 pKa = 3.88TGPDD904 pKa = 3.48GDD906 pKa = 4.85DD907 pKa = 3.17PVSFMRR913 pKa = 11.84AICRR917 pKa = 11.84DD918 pKa = 2.88SSGNVVSVTDD928 pKa = 3.76TEE930 pKa = 4.53LDD932 pKa = 3.15AVTPYY937 pKa = 9.9TVRR940 pKa = 11.84SS941 pKa = 3.62
Molecular weight: 98.27 kDa
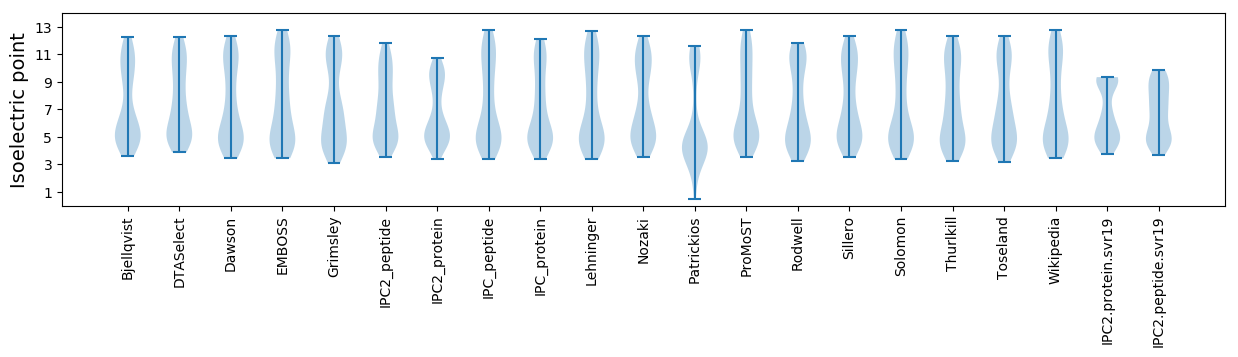
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I4AZE8|I4AZE8_9CAUD ExsB OS=Saccharomonospora phage PIS 136 OX=182851 GN=thiL PE=4 SV=1
MM1 pKa = 7.53SRR3 pKa = 11.84RR4 pKa = 11.84DD5 pKa = 3.4PHH7 pKa = 6.15TEE9 pKa = 3.55LLALEE14 pKa = 4.4LEE16 pKa = 4.26RR17 pKa = 11.84RR18 pKa = 11.84GYY20 pKa = 9.96RR21 pKa = 11.84IRR23 pKa = 11.84WRR25 pKa = 11.84HH26 pKa = 4.48NWCFTATRR34 pKa = 11.84NGRR37 pKa = 11.84SEE39 pKa = 4.69VFWSTRR45 pKa = 11.84SSLNSQIASQVAKK58 pKa = 10.53RR59 pKa = 11.84KK60 pKa = 9.71DD61 pKa = 3.01ICSRR65 pKa = 11.84LLTDD69 pKa = 4.77AGVSAPTEE77 pKa = 3.87RR78 pKa = 11.84LFRR81 pKa = 11.84ASSASAAAEE90 pKa = 4.04WASGRR95 pKa = 11.84WPVVVKK101 pKa = 9.81PATSPGHH108 pKa = 5.05STGATVGVTTADD120 pKa = 3.51GVRR123 pKa = 11.84TAVAKK128 pKa = 10.08AARR131 pKa = 11.84VSSLVLVQQQVRR143 pKa = 11.84GSEE146 pKa = 3.58ARR148 pKa = 11.84MLVVGGRR155 pKa = 11.84CVHH158 pKa = 6.07VLRR161 pKa = 11.84KK162 pKa = 9.61LGKK165 pKa = 8.01PGAYY169 pKa = 9.18EE170 pKa = 4.87AITEE174 pKa = 4.28RR175 pKa = 11.84VHH177 pKa = 6.97PSYY180 pKa = 11.08LRR182 pKa = 11.84AAEE185 pKa = 4.08RR186 pKa = 11.84AVAAIPGLGLAGLDD200 pKa = 3.72VIAEE204 pKa = 4.24DD205 pKa = 3.09WHH207 pKa = 7.63KK208 pKa = 9.9PGKK211 pKa = 9.68YY212 pKa = 9.9AVIEE216 pKa = 4.34VNSAPGIKK224 pKa = 9.59GHH226 pKa = 6.5HH227 pKa = 6.7RR228 pKa = 11.84PTKK231 pKa = 10.54GRR233 pKa = 11.84GFDD236 pKa = 3.22AAGAIVDD243 pKa = 3.69EE244 pKa = 4.76LEE246 pKa = 3.94RR247 pKa = 11.84RR248 pKa = 11.84TT249 pKa = 3.76
MM1 pKa = 7.53SRR3 pKa = 11.84RR4 pKa = 11.84DD5 pKa = 3.4PHH7 pKa = 6.15TEE9 pKa = 3.55LLALEE14 pKa = 4.4LEE16 pKa = 4.26RR17 pKa = 11.84RR18 pKa = 11.84GYY20 pKa = 9.96RR21 pKa = 11.84IRR23 pKa = 11.84WRR25 pKa = 11.84HH26 pKa = 4.48NWCFTATRR34 pKa = 11.84NGRR37 pKa = 11.84SEE39 pKa = 4.69VFWSTRR45 pKa = 11.84SSLNSQIASQVAKK58 pKa = 10.53RR59 pKa = 11.84KK60 pKa = 9.71DD61 pKa = 3.01ICSRR65 pKa = 11.84LLTDD69 pKa = 4.77AGVSAPTEE77 pKa = 3.87RR78 pKa = 11.84LFRR81 pKa = 11.84ASSASAAAEE90 pKa = 4.04WASGRR95 pKa = 11.84WPVVVKK101 pKa = 9.81PATSPGHH108 pKa = 5.05STGATVGVTTADD120 pKa = 3.51GVRR123 pKa = 11.84TAVAKK128 pKa = 10.08AARR131 pKa = 11.84VSSLVLVQQQVRR143 pKa = 11.84GSEE146 pKa = 3.58ARR148 pKa = 11.84MLVVGGRR155 pKa = 11.84CVHH158 pKa = 6.07VLRR161 pKa = 11.84KK162 pKa = 9.61LGKK165 pKa = 8.01PGAYY169 pKa = 9.18EE170 pKa = 4.87AITEE174 pKa = 4.28RR175 pKa = 11.84VHH177 pKa = 6.97PSYY180 pKa = 11.08LRR182 pKa = 11.84AAEE185 pKa = 4.08RR186 pKa = 11.84AVAAIPGLGLAGLDD200 pKa = 3.72VIAEE204 pKa = 4.24DD205 pKa = 3.09WHH207 pKa = 7.63KK208 pKa = 9.9PGKK211 pKa = 9.68YY212 pKa = 9.9AVIEE216 pKa = 4.34VNSAPGIKK224 pKa = 9.59GHH226 pKa = 6.5HH227 pKa = 6.7RR228 pKa = 11.84PTKK231 pKa = 10.54GRR233 pKa = 11.84GFDD236 pKa = 3.22AAGAIVDD243 pKa = 3.69EE244 pKa = 4.76LEE246 pKa = 3.94RR247 pKa = 11.84RR248 pKa = 11.84TT249 pKa = 3.76
Molecular weight: 26.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
28532 |
34 |
1238 |
216.2 |
23.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.138 ± 0.419 | 1.535 ± 0.162 |
6.575 ± 0.241 | 5.822 ± 0.232 |
2.324 ± 0.122 | 8.065 ± 0.281 |
2.87 ± 0.184 | 3.491 ± 0.139 |
2.555 ± 0.197 | 8.363 ± 0.313 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.11 ± 0.138 | 2.699 ± 0.138 |
6.046 ± 0.229 | 3.603 ± 0.184 |
8.825 ± 0.44 | 5.66 ± 0.345 |
6.677 ± 0.259 | 8.086 ± 0.275 |
1.77 ± 0.111 | 1.787 ± 0.124 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
