
Dishui lake virophage 1
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Maveriviricetes; Priklausovirales; Lavidaviridae; unclassified Lavidaviridae
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
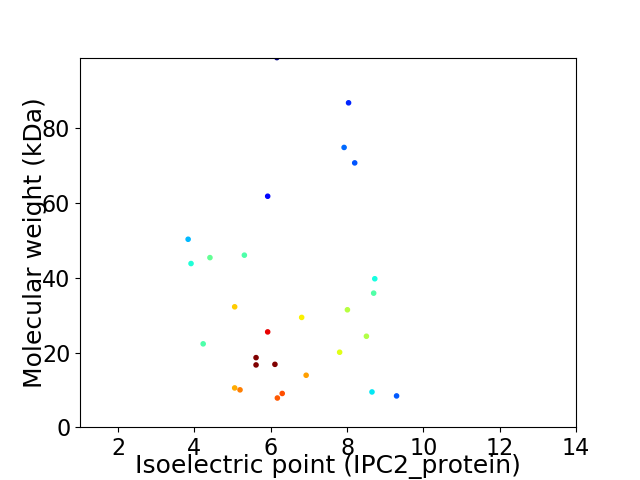
Virtual 2D-PAGE plot for 28 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A141HRM6|A0A141HRM6_9VIRU Uncharacterized protein OS=Dishui lake virophage 1 OX=1739971 PE=4 SV=1
MM1 pKa = 7.24SVSSILNPATGKK13 pKa = 10.33IYY15 pKa = 10.61DD16 pKa = 4.26DD17 pKa = 4.94LVPQGGGVPLAKK29 pKa = 10.36GGLITANAGGAEE41 pKa = 3.98QALAVGNDD49 pKa = 3.12GWVLSADD56 pKa = 3.74SNEE59 pKa = 3.98ALGLKK64 pKa = 9.55YY65 pKa = 10.16IQLPAGGIQFTQQGQLLYY83 pKa = 10.68AGAGPAFNDD92 pKa = 3.3SLLNIGNAGQILGINAGIPAWINAGGSGTITALAPLTEE130 pKa = 4.45YY131 pKa = 11.54ADD133 pKa = 3.79GTASKK138 pKa = 10.69VAVNFTGKK146 pKa = 10.69GDD148 pKa = 4.61LIVGGGVQVGGEE160 pKa = 4.03PVAGVILPVGANDD173 pKa = 3.57MVLTANSATASGLEE187 pKa = 4.12WKK189 pKa = 10.59ASGGGSSATIFRR201 pKa = 11.84NSVDD205 pKa = 3.72DD206 pKa = 3.94TPLVITKK213 pKa = 9.1PATANDD219 pKa = 3.93TCVITADD226 pKa = 3.99RR227 pKa = 11.84TYY229 pKa = 10.78HH230 pKa = 7.0AYY232 pKa = 8.06NTQIYY237 pKa = 9.45SKK239 pKa = 10.37GGVGTQVDD247 pKa = 4.09MTNPQPFPFFTWTAPNDD264 pKa = 3.38INMTSVSSQVKK275 pKa = 9.99IITNSIYY282 pKa = 10.65GQTVHH287 pKa = 7.89LDD289 pKa = 3.3IYY291 pKa = 10.97DD292 pKa = 3.81STGTTGLVGGDD303 pKa = 3.66PVNVSDD309 pKa = 5.68NEE311 pKa = 4.15PAGDD315 pKa = 3.82AVSLPITTAFQFVNGTTYY333 pKa = 11.16QFMVSLDD340 pKa = 3.69GLDD343 pKa = 3.16QGEE346 pKa = 4.35FFDD349 pKa = 4.19MFIDD353 pKa = 4.56GGQYY357 pKa = 10.2VGNITINGVDD367 pKa = 4.11YY368 pKa = 11.45ANLPATFTLAPPYY381 pKa = 10.06KK382 pKa = 10.26FRR384 pKa = 11.84VSNDD388 pKa = 3.18LTGKK392 pKa = 8.94TSATCEE398 pKa = 4.13NFSSQSFVASEE409 pKa = 4.52DD410 pKa = 3.83TNSWIAVGGINAGVAFVPP428 pKa = 4.51
MM1 pKa = 7.24SVSSILNPATGKK13 pKa = 10.33IYY15 pKa = 10.61DD16 pKa = 4.26DD17 pKa = 4.94LVPQGGGVPLAKK29 pKa = 10.36GGLITANAGGAEE41 pKa = 3.98QALAVGNDD49 pKa = 3.12GWVLSADD56 pKa = 3.74SNEE59 pKa = 3.98ALGLKK64 pKa = 9.55YY65 pKa = 10.16IQLPAGGIQFTQQGQLLYY83 pKa = 10.68AGAGPAFNDD92 pKa = 3.3SLLNIGNAGQILGINAGIPAWINAGGSGTITALAPLTEE130 pKa = 4.45YY131 pKa = 11.54ADD133 pKa = 3.79GTASKK138 pKa = 10.69VAVNFTGKK146 pKa = 10.69GDD148 pKa = 4.61LIVGGGVQVGGEE160 pKa = 4.03PVAGVILPVGANDD173 pKa = 3.57MVLTANSATASGLEE187 pKa = 4.12WKK189 pKa = 10.59ASGGGSSATIFRR201 pKa = 11.84NSVDD205 pKa = 3.72DD206 pKa = 3.94TPLVITKK213 pKa = 9.1PATANDD219 pKa = 3.93TCVITADD226 pKa = 3.99RR227 pKa = 11.84TYY229 pKa = 10.78HH230 pKa = 7.0AYY232 pKa = 8.06NTQIYY237 pKa = 9.45SKK239 pKa = 10.37GGVGTQVDD247 pKa = 4.09MTNPQPFPFFTWTAPNDD264 pKa = 3.38INMTSVSSQVKK275 pKa = 9.99IITNSIYY282 pKa = 10.65GQTVHH287 pKa = 7.89LDD289 pKa = 3.3IYY291 pKa = 10.97DD292 pKa = 3.81STGTTGLVGGDD303 pKa = 3.66PVNVSDD309 pKa = 5.68NEE311 pKa = 4.15PAGDD315 pKa = 3.82AVSLPITTAFQFVNGTTYY333 pKa = 11.16QFMVSLDD340 pKa = 3.69GLDD343 pKa = 3.16QGEE346 pKa = 4.35FFDD349 pKa = 4.19MFIDD353 pKa = 4.56GGQYY357 pKa = 10.2VGNITINGVDD367 pKa = 4.11YY368 pKa = 11.45ANLPATFTLAPPYY381 pKa = 10.06KK382 pKa = 10.26FRR384 pKa = 11.84VSNDD388 pKa = 3.18LTGKK392 pKa = 8.94TSATCEE398 pKa = 4.13NFSSQSFVASEE409 pKa = 4.52DD410 pKa = 3.83TNSWIAVGGINAGVAFVPP428 pKa = 4.51
Molecular weight: 43.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A141HRN6|A0A141HRN6_9VIRU Uncharacterized protein OS=Dishui lake virophage 1 OX=1739971 PE=4 SV=1
MM1 pKa = 7.32LCSYY5 pKa = 10.27SKK7 pKa = 10.95KK8 pKa = 10.46SFSATLPSRR17 pKa = 11.84DD18 pKa = 3.35FSTASFNSSTLRR30 pKa = 11.84SITDD34 pKa = 3.36YY35 pKa = 10.98CSSARR40 pKa = 11.84FLIISLNASSCVGSAFRR57 pKa = 11.84FFQNLIHH64 pKa = 6.85SGRR67 pKa = 11.84KK68 pKa = 6.59YY69 pKa = 10.55HH70 pKa = 6.99ISLFII75 pKa = 4.92
MM1 pKa = 7.32LCSYY5 pKa = 10.27SKK7 pKa = 10.95KK8 pKa = 10.46SFSATLPSRR17 pKa = 11.84DD18 pKa = 3.35FSTASFNSSTLRR30 pKa = 11.84SITDD34 pKa = 3.36YY35 pKa = 10.98CSSARR40 pKa = 11.84FLIISLNASSCVGSAFRR57 pKa = 11.84FFQNLIHH64 pKa = 6.85SGRR67 pKa = 11.84KK68 pKa = 6.59YY69 pKa = 10.55HH70 pKa = 6.99ISLFII75 pKa = 4.92
Molecular weight: 8.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8703 |
63 |
857 |
310.8 |
34.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.836 ± 0.412 | 1.126 ± 0.189 |
5.538 ± 0.355 | 5.872 ± 0.685 |
3.976 ± 0.214 | 8.307 ± 1.191 |
1.241 ± 0.185 | 5.607 ± 0.376 |
6.71 ± 0.731 | 8.009 ± 0.345 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.873 ± 0.214 | 5.676 ± 0.253 |
4.998 ± 0.467 | 4.929 ± 0.491 |
4.309 ± 0.548 | 7.055 ± 0.592 |
6.182 ± 0.685 | 5.607 ± 0.375 |
0.954 ± 0.121 | 4.194 ± 0.316 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
