
Colletotrichum fructicola chrysovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Chrysoviridae; Betachrysovirus
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins
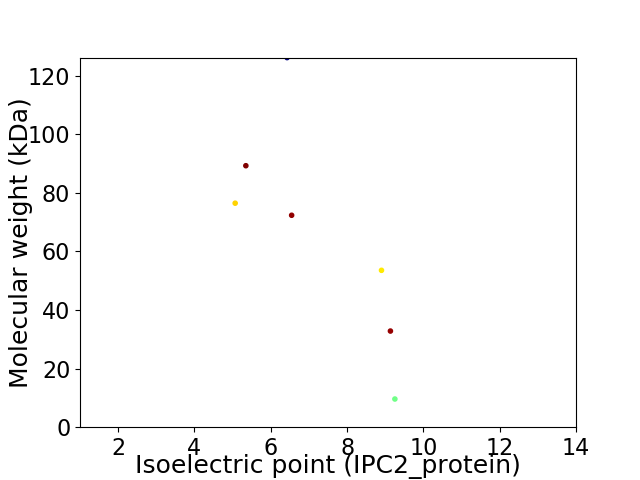
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346IME2|A0A346IME2_9VIRU Uncharacterized protein OS=Colletotrichum fructicola chrysovirus 1 OX=2304034 PE=4 SV=1
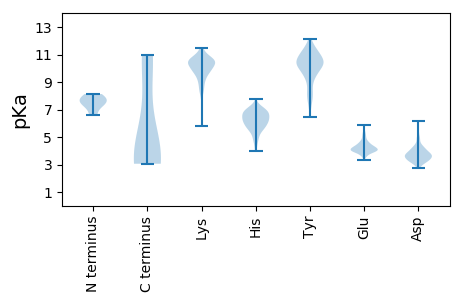
MM1 pKa = 6.59STPYY5 pKa = 10.42FGVNIDD11 pKa = 3.56ARR13 pKa = 11.84LMEE16 pKa = 5.71LDD18 pKa = 3.56DD19 pKa = 4.4AYY21 pKa = 10.24KK22 pKa = 10.23FRR24 pKa = 11.84RR25 pKa = 11.84ISARR29 pKa = 11.84QLIEE33 pKa = 3.96HH34 pKa = 6.26VEE36 pKa = 3.91PLLPLLEE43 pKa = 4.64GPVPQEE49 pKa = 3.7TRR51 pKa = 11.84DD52 pKa = 3.58SVRR55 pKa = 11.84NAEE58 pKa = 4.13RR59 pKa = 11.84EE60 pKa = 4.11LGEE63 pKa = 3.92VTRR66 pKa = 11.84LHH68 pKa = 6.25EE69 pKa = 5.25CGLGTADD76 pKa = 4.38CATEE80 pKa = 3.89RR81 pKa = 11.84GGGEE85 pKa = 3.94EE86 pKa = 4.31LVVEE90 pKa = 4.53VARR93 pKa = 11.84VYY95 pKa = 11.36SDD97 pKa = 3.3AAAAATKK104 pKa = 9.86HH105 pKa = 6.41HH106 pKa = 6.71NEE108 pKa = 3.5QSEE111 pKa = 3.99WGRR114 pKa = 11.84NRR116 pKa = 11.84DD117 pKa = 3.19TRR119 pKa = 11.84IMRR122 pKa = 11.84RR123 pKa = 11.84GHH125 pKa = 6.77HH126 pKa = 7.05DD127 pKa = 3.14VTPINRR133 pKa = 11.84NLISMSYY140 pKa = 10.06GLPKK144 pKa = 10.53GADD147 pKa = 3.24VRR149 pKa = 11.84ALAWTDD155 pKa = 3.1ADD157 pKa = 3.6VSRR160 pKa = 11.84PRR162 pKa = 11.84LLNTLVAGASSLHH175 pKa = 5.91KK176 pKa = 10.29LWRR179 pKa = 11.84LQACSVTLPSGRR191 pKa = 11.84VLMSEE196 pKa = 4.93AYY198 pKa = 9.17PGRR201 pKa = 11.84GLGGMDD207 pKa = 2.83MYY209 pKa = 11.24GFNLHH214 pKa = 6.36EE215 pKa = 4.31VALYY219 pKa = 8.27LTISEE224 pKa = 4.76DD225 pKa = 3.61LLVDD229 pKa = 3.37VSVAGKK235 pKa = 9.82PEE237 pKa = 3.8GLRR240 pKa = 11.84VTNRR244 pKa = 11.84EE245 pKa = 3.94RR246 pKa = 11.84FDD248 pKa = 3.76EE249 pKa = 4.91FLDD252 pKa = 3.83LCSIVQFWIVDD263 pKa = 3.39RR264 pKa = 11.84MVPISGLNVLEE275 pKa = 5.2GYY277 pKa = 8.2TLAQLKK283 pKa = 8.45TLTTRR288 pKa = 11.84TQQACTPSGHH298 pKa = 5.62MVRR301 pKa = 11.84GPIGALVSLGEE312 pKa = 4.1WEE314 pKa = 4.76ADD316 pKa = 3.37DD317 pKa = 4.11SEE319 pKa = 5.52KK320 pKa = 10.76IGVSHH325 pKa = 7.05CAADD329 pKa = 3.98GTVSRR334 pKa = 11.84GTLSPNNIPPGLLTVYY350 pKa = 8.78ATSGGKK356 pKa = 7.59TVTGGTLTLRR366 pKa = 11.84GEE368 pKa = 4.23EE369 pKa = 4.07GVYY372 pKa = 9.73TVTSSVTANMTIGHH386 pKa = 6.35IKK388 pKa = 9.92EE389 pKa = 4.21LSKK392 pKa = 10.84CAPTVRR398 pKa = 11.84DD399 pKa = 3.62GLFSPTAAGALLTDD413 pKa = 4.22GDD415 pKa = 3.86GGIYY419 pKa = 10.15VDD421 pKa = 4.18VNHH424 pKa = 6.28EE425 pKa = 4.24HH426 pKa = 6.26VVVPTIFSGVPDD438 pKa = 3.92GSEE441 pKa = 3.69IEE443 pKa = 4.46VVARR447 pKa = 11.84NCSAMLGYY455 pKa = 10.64LEE457 pKa = 5.86DD458 pKa = 3.69VAALQGWSYY467 pKa = 11.3SVEE470 pKa = 3.94LGGTQFNIDD479 pKa = 3.87PSEE482 pKa = 4.42DD483 pKa = 3.73GAWARR488 pKa = 11.84IVVKK492 pKa = 10.34IRR494 pKa = 11.84YY495 pKa = 8.28GPEE498 pKa = 3.54VRR500 pKa = 11.84PQWATIVAGEE510 pKa = 4.14LDD512 pKa = 3.95EE513 pKa = 5.32LAVDD517 pKa = 3.59SEE519 pKa = 4.21LRR521 pKa = 11.84LDD523 pKa = 3.83YY524 pKa = 10.86GVSGLGRR531 pKa = 11.84YY532 pKa = 8.64LLSRR536 pKa = 11.84FLVGGTFIPGTSLKK550 pKa = 10.07INKK553 pKa = 9.3RR554 pKa = 11.84GRR556 pKa = 11.84FGAIHH561 pKa = 6.75GVLKK565 pKa = 10.29EE566 pKa = 4.03NKK568 pKa = 8.98AALMLTGSLPTPTNYY583 pKa = 9.71VVDD586 pKa = 4.24VKK588 pKa = 10.36TLARR592 pKa = 11.84TRR594 pKa = 11.84GSEE597 pKa = 3.99TQYY600 pKa = 9.26YY601 pKa = 10.0AYY603 pKa = 10.27AIGVAKK609 pKa = 9.71FIRR612 pKa = 11.84NQYY615 pKa = 9.16VGSACCVDD623 pKa = 4.93DD624 pKa = 5.62SPEE627 pKa = 4.02LDD629 pKa = 3.55EE630 pKa = 5.44FLSANRR636 pKa = 11.84DD637 pKa = 3.66EE638 pKa = 5.82LPDD641 pKa = 3.58EE642 pKa = 4.71VGALLANPEE651 pKa = 4.21VPSRR655 pKa = 11.84DD656 pKa = 3.27VTTVLSEE663 pKa = 3.97LRR665 pKa = 11.84ALEE668 pKa = 4.26SPPTCVCSLRR678 pKa = 11.84DD679 pKa = 3.34VDD681 pKa = 5.37TSTQCYY687 pKa = 7.94MEE689 pKa = 4.77SMSAGPVYY697 pKa = 10.69SGGRR701 pKa = 11.84IITVLYY707 pKa = 10.63
MM1 pKa = 6.59STPYY5 pKa = 10.42FGVNIDD11 pKa = 3.56ARR13 pKa = 11.84LMEE16 pKa = 5.71LDD18 pKa = 3.56DD19 pKa = 4.4AYY21 pKa = 10.24KK22 pKa = 10.23FRR24 pKa = 11.84RR25 pKa = 11.84ISARR29 pKa = 11.84QLIEE33 pKa = 3.96HH34 pKa = 6.26VEE36 pKa = 3.91PLLPLLEE43 pKa = 4.64GPVPQEE49 pKa = 3.7TRR51 pKa = 11.84DD52 pKa = 3.58SVRR55 pKa = 11.84NAEE58 pKa = 4.13RR59 pKa = 11.84EE60 pKa = 4.11LGEE63 pKa = 3.92VTRR66 pKa = 11.84LHH68 pKa = 6.25EE69 pKa = 5.25CGLGTADD76 pKa = 4.38CATEE80 pKa = 3.89RR81 pKa = 11.84GGGEE85 pKa = 3.94EE86 pKa = 4.31LVVEE90 pKa = 4.53VARR93 pKa = 11.84VYY95 pKa = 11.36SDD97 pKa = 3.3AAAAATKK104 pKa = 9.86HH105 pKa = 6.41HH106 pKa = 6.71NEE108 pKa = 3.5QSEE111 pKa = 3.99WGRR114 pKa = 11.84NRR116 pKa = 11.84DD117 pKa = 3.19TRR119 pKa = 11.84IMRR122 pKa = 11.84RR123 pKa = 11.84GHH125 pKa = 6.77HH126 pKa = 7.05DD127 pKa = 3.14VTPINRR133 pKa = 11.84NLISMSYY140 pKa = 10.06GLPKK144 pKa = 10.53GADD147 pKa = 3.24VRR149 pKa = 11.84ALAWTDD155 pKa = 3.1ADD157 pKa = 3.6VSRR160 pKa = 11.84PRR162 pKa = 11.84LLNTLVAGASSLHH175 pKa = 5.91KK176 pKa = 10.29LWRR179 pKa = 11.84LQACSVTLPSGRR191 pKa = 11.84VLMSEE196 pKa = 4.93AYY198 pKa = 9.17PGRR201 pKa = 11.84GLGGMDD207 pKa = 2.83MYY209 pKa = 11.24GFNLHH214 pKa = 6.36EE215 pKa = 4.31VALYY219 pKa = 8.27LTISEE224 pKa = 4.76DD225 pKa = 3.61LLVDD229 pKa = 3.37VSVAGKK235 pKa = 9.82PEE237 pKa = 3.8GLRR240 pKa = 11.84VTNRR244 pKa = 11.84EE245 pKa = 3.94RR246 pKa = 11.84FDD248 pKa = 3.76EE249 pKa = 4.91FLDD252 pKa = 3.83LCSIVQFWIVDD263 pKa = 3.39RR264 pKa = 11.84MVPISGLNVLEE275 pKa = 5.2GYY277 pKa = 8.2TLAQLKK283 pKa = 8.45TLTTRR288 pKa = 11.84TQQACTPSGHH298 pKa = 5.62MVRR301 pKa = 11.84GPIGALVSLGEE312 pKa = 4.1WEE314 pKa = 4.76ADD316 pKa = 3.37DD317 pKa = 4.11SEE319 pKa = 5.52KK320 pKa = 10.76IGVSHH325 pKa = 7.05CAADD329 pKa = 3.98GTVSRR334 pKa = 11.84GTLSPNNIPPGLLTVYY350 pKa = 8.78ATSGGKK356 pKa = 7.59TVTGGTLTLRR366 pKa = 11.84GEE368 pKa = 4.23EE369 pKa = 4.07GVYY372 pKa = 9.73TVTSSVTANMTIGHH386 pKa = 6.35IKK388 pKa = 9.92EE389 pKa = 4.21LSKK392 pKa = 10.84CAPTVRR398 pKa = 11.84DD399 pKa = 3.62GLFSPTAAGALLTDD413 pKa = 4.22GDD415 pKa = 3.86GGIYY419 pKa = 10.15VDD421 pKa = 4.18VNHH424 pKa = 6.28EE425 pKa = 4.24HH426 pKa = 6.26VVVPTIFSGVPDD438 pKa = 3.92GSEE441 pKa = 3.69IEE443 pKa = 4.46VVARR447 pKa = 11.84NCSAMLGYY455 pKa = 10.64LEE457 pKa = 5.86DD458 pKa = 3.69VAALQGWSYY467 pKa = 11.3SVEE470 pKa = 3.94LGGTQFNIDD479 pKa = 3.87PSEE482 pKa = 4.42DD483 pKa = 3.73GAWARR488 pKa = 11.84IVVKK492 pKa = 10.34IRR494 pKa = 11.84YY495 pKa = 8.28GPEE498 pKa = 3.54VRR500 pKa = 11.84PQWATIVAGEE510 pKa = 4.14LDD512 pKa = 3.95EE513 pKa = 5.32LAVDD517 pKa = 3.59SEE519 pKa = 4.21LRR521 pKa = 11.84LDD523 pKa = 3.83YY524 pKa = 10.86GVSGLGRR531 pKa = 11.84YY532 pKa = 8.64LLSRR536 pKa = 11.84FLVGGTFIPGTSLKK550 pKa = 10.07INKK553 pKa = 9.3RR554 pKa = 11.84GRR556 pKa = 11.84FGAIHH561 pKa = 6.75GVLKK565 pKa = 10.29EE566 pKa = 4.03NKK568 pKa = 8.98AALMLTGSLPTPTNYY583 pKa = 9.71VVDD586 pKa = 4.24VKK588 pKa = 10.36TLARR592 pKa = 11.84TRR594 pKa = 11.84GSEE597 pKa = 3.99TQYY600 pKa = 9.26YY601 pKa = 10.0AYY603 pKa = 10.27AIGVAKK609 pKa = 9.71FIRR612 pKa = 11.84NQYY615 pKa = 9.16VGSACCVDD623 pKa = 4.93DD624 pKa = 5.62SPEE627 pKa = 4.02LDD629 pKa = 3.55EE630 pKa = 5.44FLSANRR636 pKa = 11.84DD637 pKa = 3.66EE638 pKa = 5.82LPDD641 pKa = 3.58EE642 pKa = 4.71VGALLANPEE651 pKa = 4.21VPSRR655 pKa = 11.84DD656 pKa = 3.27VTTVLSEE663 pKa = 3.97LRR665 pKa = 11.84ALEE668 pKa = 4.26SPPTCVCSLRR678 pKa = 11.84DD679 pKa = 3.34VDD681 pKa = 5.37TSTQCYY687 pKa = 7.94MEE689 pKa = 4.77SMSAGPVYY697 pKa = 10.69SGGRR701 pKa = 11.84IITVLYY707 pKa = 10.63
Molecular weight: 76.49 kDa
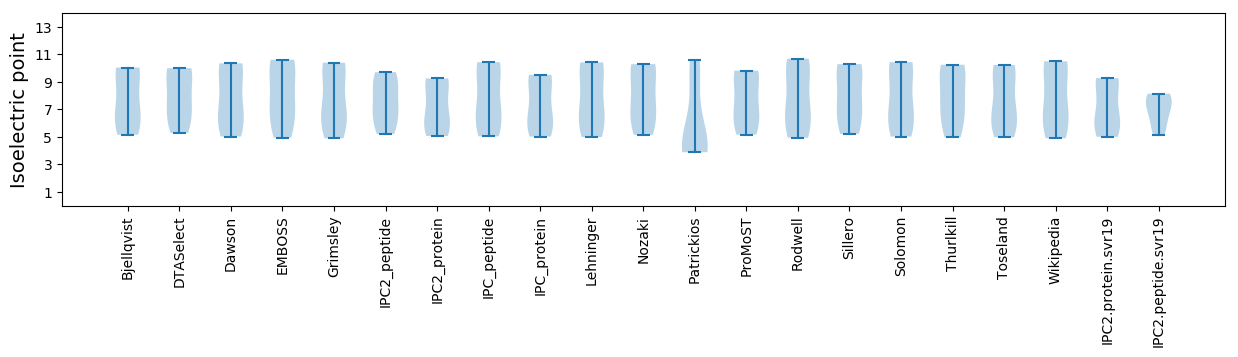
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346IME4|A0A346IME4_9VIRU Uncharacterized protein OS=Colletotrichum fructicola chrysovirus 1 OX=2304034 PE=4 SV=1
MM1 pKa = 8.12DD2 pKa = 4.38SSRR5 pKa = 11.84DD6 pKa = 3.52KK7 pKa = 11.37KK8 pKa = 10.78EE9 pKa = 3.6AGQRR13 pKa = 11.84RR14 pKa = 11.84LVSARR19 pKa = 11.84WRR21 pKa = 11.84VRR23 pKa = 11.84RR24 pKa = 11.84PTNASSEE31 pKa = 4.49TATSVSSNTSFPATVAHH48 pKa = 6.81LFYY51 pKa = 11.21VVGDD55 pKa = 3.78SFDD58 pKa = 4.15FFVGDD63 pKa = 3.86TFVPCYY69 pKa = 10.73AEE71 pKa = 4.04FVRR74 pKa = 11.84VFEE77 pKa = 4.76PGSRR81 pKa = 11.84CYY83 pKa = 11.02AWFVSPAVEE92 pKa = 3.89FRR94 pKa = 11.84FRR96 pKa = 11.84VDD98 pKa = 3.26TLPLGGRR105 pKa = 11.84RR106 pKa = 11.84AVRR109 pKa = 11.84ALRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84LSSCFSRR121 pKa = 11.84RR122 pKa = 11.84AALLASPAQLARR134 pKa = 11.84TRR136 pKa = 11.84AVRR139 pKa = 11.84FVRR142 pKa = 11.84ASEE145 pKa = 4.39GGPEE149 pKa = 4.03PKK151 pKa = 9.98SAEE154 pKa = 3.98RR155 pKa = 11.84APVCDD160 pKa = 3.42NRR162 pKa = 11.84FFSPRR167 pKa = 11.84AVEE170 pKa = 4.01SLSGFEE176 pKa = 4.06HH177 pKa = 6.54AVVGTRR183 pKa = 11.84LEE185 pKa = 4.36APASVWLRR193 pKa = 11.84EE194 pKa = 3.93EE195 pKa = 4.74GVRR198 pKa = 11.84AWGSVSDD205 pKa = 3.93LSARR209 pKa = 11.84GEE211 pKa = 3.83RR212 pKa = 11.84RR213 pKa = 11.84AALSSRR219 pKa = 11.84FLSAVTPWSPEE230 pKa = 3.66LPLQKK235 pKa = 9.71PRR237 pKa = 11.84AVVRR241 pKa = 11.84PPTSVAPLVLQAAVAAPVLRR261 pKa = 11.84TEE263 pKa = 4.06VAFDD267 pKa = 3.48SGALASVWGPRR278 pKa = 11.84RR279 pKa = 11.84EE280 pKa = 3.97EE281 pKa = 4.07SALGPGASGSGAGGFAALEE300 pKa = 4.13ASLAPSVRR308 pKa = 11.84APSGGGFAALEE319 pKa = 4.16ARR321 pKa = 11.84TAALGSSGLLGGGCGAVGSRR341 pKa = 11.84SFSSAQGTRR350 pKa = 11.84VGSLSGEE357 pKa = 4.18CATRR361 pKa = 11.84EE362 pKa = 4.07VSDD365 pKa = 3.3VTVTVEE371 pKa = 4.32PPADD375 pKa = 3.67DD376 pKa = 4.32SFTANRR382 pKa = 11.84ALPSFRR388 pKa = 11.84EE389 pKa = 4.24VVRR392 pKa = 11.84SSLGASDD399 pKa = 4.86DD400 pKa = 4.03GSGGLAAVGVPSSLRR415 pKa = 11.84RR416 pKa = 11.84RR417 pKa = 11.84RR418 pKa = 11.84GFACHH423 pKa = 6.54KK424 pKa = 10.15CGAQFSDD431 pKa = 3.54ALVLSTHH438 pKa = 6.42TSGCPNHH445 pKa = 6.93RR446 pKa = 11.84CICGKK451 pKa = 9.2PCRR454 pKa = 11.84THH456 pKa = 6.68AQRR459 pKa = 11.84QEE461 pKa = 3.91CVGRR465 pKa = 11.84HH466 pKa = 4.68FAGEE470 pKa = 4.07PFPCPVCFSGSFRR483 pKa = 11.84TFAEE487 pKa = 4.15LCAHH491 pKa = 5.8VRR493 pKa = 11.84EE494 pKa = 4.67APCRR498 pKa = 11.84RR499 pKa = 11.84LEE501 pKa = 4.27LGRR504 pKa = 11.84ASS506 pKa = 3.39
MM1 pKa = 8.12DD2 pKa = 4.38SSRR5 pKa = 11.84DD6 pKa = 3.52KK7 pKa = 11.37KK8 pKa = 10.78EE9 pKa = 3.6AGQRR13 pKa = 11.84RR14 pKa = 11.84LVSARR19 pKa = 11.84WRR21 pKa = 11.84VRR23 pKa = 11.84RR24 pKa = 11.84PTNASSEE31 pKa = 4.49TATSVSSNTSFPATVAHH48 pKa = 6.81LFYY51 pKa = 11.21VVGDD55 pKa = 3.78SFDD58 pKa = 4.15FFVGDD63 pKa = 3.86TFVPCYY69 pKa = 10.73AEE71 pKa = 4.04FVRR74 pKa = 11.84VFEE77 pKa = 4.76PGSRR81 pKa = 11.84CYY83 pKa = 11.02AWFVSPAVEE92 pKa = 3.89FRR94 pKa = 11.84FRR96 pKa = 11.84VDD98 pKa = 3.26TLPLGGRR105 pKa = 11.84RR106 pKa = 11.84AVRR109 pKa = 11.84ALRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84LSSCFSRR121 pKa = 11.84RR122 pKa = 11.84AALLASPAQLARR134 pKa = 11.84TRR136 pKa = 11.84AVRR139 pKa = 11.84FVRR142 pKa = 11.84ASEE145 pKa = 4.39GGPEE149 pKa = 4.03PKK151 pKa = 9.98SAEE154 pKa = 3.98RR155 pKa = 11.84APVCDD160 pKa = 3.42NRR162 pKa = 11.84FFSPRR167 pKa = 11.84AVEE170 pKa = 4.01SLSGFEE176 pKa = 4.06HH177 pKa = 6.54AVVGTRR183 pKa = 11.84LEE185 pKa = 4.36APASVWLRR193 pKa = 11.84EE194 pKa = 3.93EE195 pKa = 4.74GVRR198 pKa = 11.84AWGSVSDD205 pKa = 3.93LSARR209 pKa = 11.84GEE211 pKa = 3.83RR212 pKa = 11.84RR213 pKa = 11.84AALSSRR219 pKa = 11.84FLSAVTPWSPEE230 pKa = 3.66LPLQKK235 pKa = 9.71PRR237 pKa = 11.84AVVRR241 pKa = 11.84PPTSVAPLVLQAAVAAPVLRR261 pKa = 11.84TEE263 pKa = 4.06VAFDD267 pKa = 3.48SGALASVWGPRR278 pKa = 11.84RR279 pKa = 11.84EE280 pKa = 3.97EE281 pKa = 4.07SALGPGASGSGAGGFAALEE300 pKa = 4.13ASLAPSVRR308 pKa = 11.84APSGGGFAALEE319 pKa = 4.16ARR321 pKa = 11.84TAALGSSGLLGGGCGAVGSRR341 pKa = 11.84SFSSAQGTRR350 pKa = 11.84VGSLSGEE357 pKa = 4.18CATRR361 pKa = 11.84EE362 pKa = 4.07VSDD365 pKa = 3.3VTVTVEE371 pKa = 4.32PPADD375 pKa = 3.67DD376 pKa = 4.32SFTANRR382 pKa = 11.84ALPSFRR388 pKa = 11.84EE389 pKa = 4.24VVRR392 pKa = 11.84SSLGASDD399 pKa = 4.86DD400 pKa = 4.03GSGGLAAVGVPSSLRR415 pKa = 11.84RR416 pKa = 11.84RR417 pKa = 11.84RR418 pKa = 11.84GFACHH423 pKa = 6.54KK424 pKa = 10.15CGAQFSDD431 pKa = 3.54ALVLSTHH438 pKa = 6.42TSGCPNHH445 pKa = 6.93RR446 pKa = 11.84CICGKK451 pKa = 9.2PCRR454 pKa = 11.84THH456 pKa = 6.68AQRR459 pKa = 11.84QEE461 pKa = 3.91CVGRR465 pKa = 11.84HH466 pKa = 4.68FAGEE470 pKa = 4.07PFPCPVCFSGSFRR483 pKa = 11.84TFAEE487 pKa = 4.15LCAHH491 pKa = 5.8VRR493 pKa = 11.84EE494 pKa = 4.67APCRR498 pKa = 11.84RR499 pKa = 11.84LEE501 pKa = 4.27LGRR504 pKa = 11.84ASS506 pKa = 3.39
Molecular weight: 53.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4224 |
89 |
1132 |
603.4 |
65.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.559 ± 0.57 | 1.823 ± 0.262 |
4.948 ± 0.338 | 5.729 ± 0.473 |
3.291 ± 0.554 | 8.333 ± 0.644 |
2.675 ± 0.361 | 2.841 ± 0.62 |
2.936 ± 0.544 | 9.446 ± 0.498 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.32 ± 0.505 | 2.509 ± 0.33 |
5.208 ± 0.501 | 3.741 ± 0.619 |
7.434 ± 0.816 | 7.647 ± 0.834 |
5.729 ± 0.447 | 8.57 ± 0.572 |
1.231 ± 0.093 | 3.03 ± 0.403 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
