
filamentous cyanobacterium CCP5
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; unclassified Cyanobacteria
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
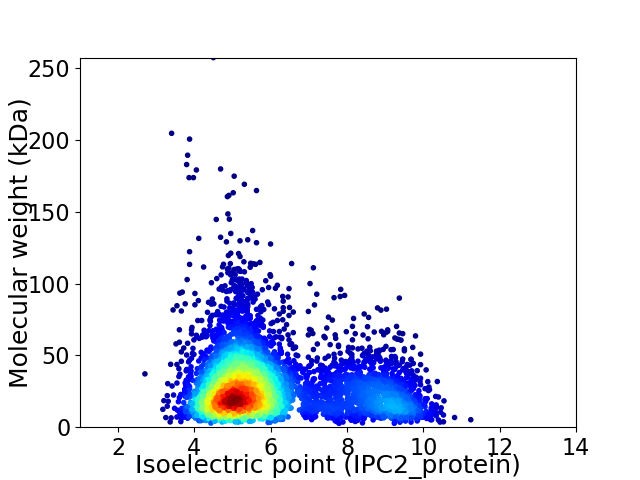
Virtual 2D-PAGE plot for 5193 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P8WHX5|A0A2P8WHX5_9CYAN S-(hydroxymethyl)glutathione dehydrogenase OS=filamentous cyanobacterium CCP5 OX=2107701 GN=C7271_07370 PE=3 SV=1
MM1 pKa = 7.46SPVILISACLLAGGCQPQSEE21 pKa = 4.76PNVDD25 pKa = 3.38APLSGEE31 pKa = 3.99QVTILGTLTGVAEE44 pKa = 4.6EE45 pKa = 4.26KK46 pKa = 11.02LEE48 pKa = 4.16AALEE52 pKa = 4.16PFTAKK57 pKa = 9.87TGIEE61 pKa = 4.12VVYY64 pKa = 10.22EE65 pKa = 4.05GTDD68 pKa = 2.95AFTTLIPVRR77 pKa = 11.84VDD79 pKa = 3.37SNNTPDD85 pKa = 3.55IALFPQPGLMADD97 pKa = 4.78FAAEE101 pKa = 4.09GQMVPLDD108 pKa = 3.79SFMDD112 pKa = 3.75MDD114 pKa = 4.24QLAAAYY120 pKa = 10.05DD121 pKa = 4.25DD122 pKa = 3.9YY123 pKa = 11.22WLEE126 pKa = 4.02LTSLDD131 pKa = 3.44GHH133 pKa = 7.02PYY135 pKa = 9.85GVWIRR140 pKa = 11.84ADD142 pKa = 3.47VKK144 pKa = 11.18SLVWYY149 pKa = 10.36NPAAFAAAGYY159 pKa = 8.49GVPTDD164 pKa = 3.69WQGLEE169 pKa = 3.96ALMDD173 pKa = 3.85QIVADD178 pKa = 5.54GGTPWCLGMEE188 pKa = 4.34SGKK191 pKa = 9.2ATGWVGTDD199 pKa = 2.85WVEE202 pKa = 3.88EE203 pKa = 3.99LLLRR207 pKa = 11.84MAGPEE212 pKa = 4.02VYY214 pKa = 10.27DD215 pKa = 2.92RR216 pKa = 11.84WVAHH220 pKa = 7.02EE221 pKa = 4.51IPFNAPEE228 pKa = 3.88VKK230 pKa = 10.32AAFEE234 pKa = 4.02QFGAIARR241 pKa = 11.84SPDD244 pKa = 3.65YY245 pKa = 10.97VYY247 pKa = 11.26GGATSAISTPFGDD260 pKa = 4.03APRR263 pKa = 11.84PLFDD267 pKa = 4.74QPPGCYY273 pKa = 9.03LHH275 pKa = 6.86RR276 pKa = 11.84QASFIEE282 pKa = 4.35EE283 pKa = 4.43FFPASAVPAEE293 pKa = 4.39TVSLFPLPAMDD304 pKa = 4.86EE305 pKa = 4.46GPAPVLVSGIVFGLFNDD322 pKa = 4.39TPAARR327 pKa = 11.84ALMEE331 pKa = 4.1YY332 pKa = 10.4LATPEE337 pKa = 4.15PHH339 pKa = 6.93EE340 pKa = 4.06IWAGLGSYY348 pKa = 10.18ISPHH352 pKa = 5.7RR353 pKa = 11.84QVGLDD358 pKa = 3.85AYY360 pKa = 10.02PDD362 pKa = 3.7VLTQRR367 pKa = 11.84QAEE370 pKa = 4.12ILQNAEE376 pKa = 3.87VVRR379 pKa = 11.84FDD381 pKa = 5.41GSDD384 pKa = 3.47LMPGAVGTGSFWSGVVDD401 pKa = 3.6YY402 pKa = 11.61VGGEE406 pKa = 4.25DD407 pKa = 4.68LDD409 pKa = 3.89QVLADD414 pKa = 5.38IEE416 pKa = 4.67ASWPTEE422 pKa = 3.8EE423 pKa = 4.69
MM1 pKa = 7.46SPVILISACLLAGGCQPQSEE21 pKa = 4.76PNVDD25 pKa = 3.38APLSGEE31 pKa = 3.99QVTILGTLTGVAEE44 pKa = 4.6EE45 pKa = 4.26KK46 pKa = 11.02LEE48 pKa = 4.16AALEE52 pKa = 4.16PFTAKK57 pKa = 9.87TGIEE61 pKa = 4.12VVYY64 pKa = 10.22EE65 pKa = 4.05GTDD68 pKa = 2.95AFTTLIPVRR77 pKa = 11.84VDD79 pKa = 3.37SNNTPDD85 pKa = 3.55IALFPQPGLMADD97 pKa = 4.78FAAEE101 pKa = 4.09GQMVPLDD108 pKa = 3.79SFMDD112 pKa = 3.75MDD114 pKa = 4.24QLAAAYY120 pKa = 10.05DD121 pKa = 4.25DD122 pKa = 3.9YY123 pKa = 11.22WLEE126 pKa = 4.02LTSLDD131 pKa = 3.44GHH133 pKa = 7.02PYY135 pKa = 9.85GVWIRR140 pKa = 11.84ADD142 pKa = 3.47VKK144 pKa = 11.18SLVWYY149 pKa = 10.36NPAAFAAAGYY159 pKa = 8.49GVPTDD164 pKa = 3.69WQGLEE169 pKa = 3.96ALMDD173 pKa = 3.85QIVADD178 pKa = 5.54GGTPWCLGMEE188 pKa = 4.34SGKK191 pKa = 9.2ATGWVGTDD199 pKa = 2.85WVEE202 pKa = 3.88EE203 pKa = 3.99LLLRR207 pKa = 11.84MAGPEE212 pKa = 4.02VYY214 pKa = 10.27DD215 pKa = 2.92RR216 pKa = 11.84WVAHH220 pKa = 7.02EE221 pKa = 4.51IPFNAPEE228 pKa = 3.88VKK230 pKa = 10.32AAFEE234 pKa = 4.02QFGAIARR241 pKa = 11.84SPDD244 pKa = 3.65YY245 pKa = 10.97VYY247 pKa = 11.26GGATSAISTPFGDD260 pKa = 4.03APRR263 pKa = 11.84PLFDD267 pKa = 4.74QPPGCYY273 pKa = 9.03LHH275 pKa = 6.86RR276 pKa = 11.84QASFIEE282 pKa = 4.35EE283 pKa = 4.43FFPASAVPAEE293 pKa = 4.39TVSLFPLPAMDD304 pKa = 4.86EE305 pKa = 4.46GPAPVLVSGIVFGLFNDD322 pKa = 4.39TPAARR327 pKa = 11.84ALMEE331 pKa = 4.1YY332 pKa = 10.4LATPEE337 pKa = 4.15PHH339 pKa = 6.93EE340 pKa = 4.06IWAGLGSYY348 pKa = 10.18ISPHH352 pKa = 5.7RR353 pKa = 11.84QVGLDD358 pKa = 3.85AYY360 pKa = 10.02PDD362 pKa = 3.7VLTQRR367 pKa = 11.84QAEE370 pKa = 4.12ILQNAEE376 pKa = 3.87VVRR379 pKa = 11.84FDD381 pKa = 5.41GSDD384 pKa = 3.47LMPGAVGTGSFWSGVVDD401 pKa = 3.6YY402 pKa = 11.61VGGEE406 pKa = 4.25DD407 pKa = 4.68LDD409 pKa = 3.89QVLADD414 pKa = 5.38IEE416 pKa = 4.67ASWPTEE422 pKa = 3.8EE423 pKa = 4.69
Molecular weight: 45.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P8WCE6|A0A2P8WCE6_9CYAN DNA-binding response regulator OS=filamentous cyanobacterium CCP5 OX=2107701 GN=C7271_17785 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 9.06RR4 pKa = 11.84TLGGTNRR11 pKa = 11.84KK12 pKa = 7.61QKK14 pKa = 8.99RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84SHH26 pKa = 7.41TGQQVIKK33 pKa = 9.99SRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.22GRR40 pKa = 11.84ARR42 pKa = 11.84LAVV45 pKa = 3.42
MM1 pKa = 7.74SKK3 pKa = 9.06RR4 pKa = 11.84TLGGTNRR11 pKa = 11.84KK12 pKa = 7.61QKK14 pKa = 8.99RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84SHH26 pKa = 7.41TGQQVIKK33 pKa = 9.99SRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.22GRR40 pKa = 11.84ARR42 pKa = 11.84LAVV45 pKa = 3.42
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1436203 |
27 |
2385 |
276.6 |
30.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.869 ± 0.04 | 0.935 ± 0.011 |
5.472 ± 0.029 | 5.869 ± 0.035 |
3.661 ± 0.024 | 7.479 ± 0.037 |
2.01 ± 0.021 | 5.549 ± 0.025 |
3.014 ± 0.025 | 11.256 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.022 ± 0.017 | 3.09 ± 0.023 |
5.354 ± 0.028 | 5.473 ± 0.036 |
6.056 ± 0.029 | 6.16 ± 0.029 |
5.417 ± 0.025 | 6.943 ± 0.029 |
1.581 ± 0.018 | 2.792 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
