
Flavobacterium phage FLiP
Taxonomy: Viruses; Finnlakeviridae; Finnlakevirus; Flavobacterium virus FLiP
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
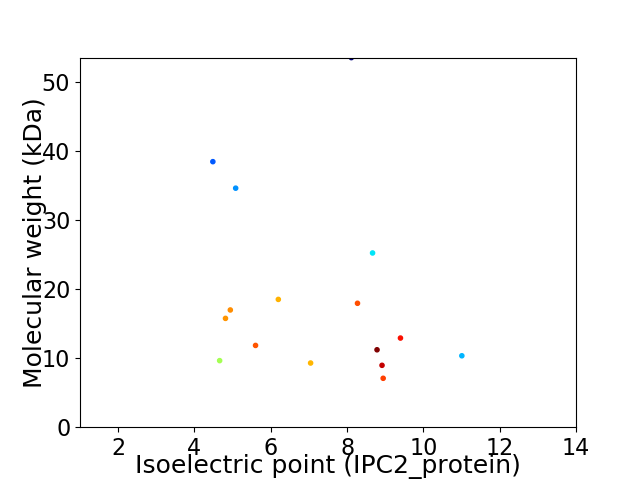
Virtual 2D-PAGE plot for 16 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
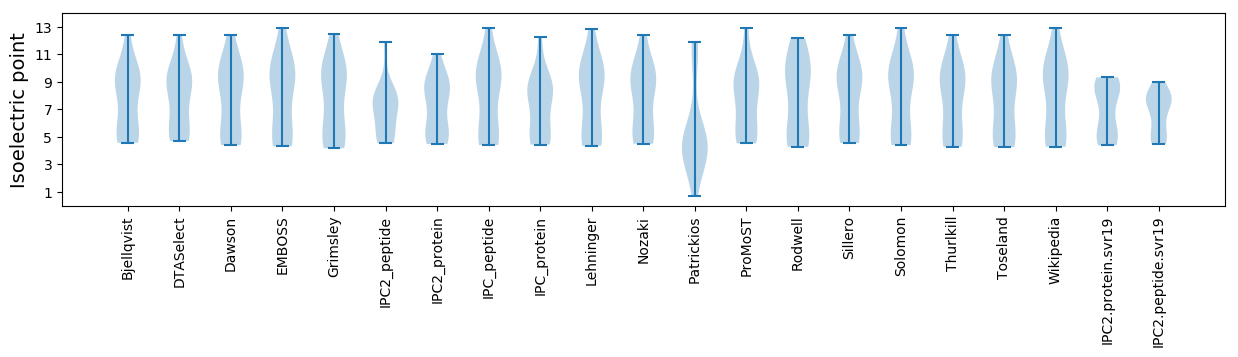
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A222NP85|A0A222NP85_9VIRU Uncharacterized protein OS=Flavobacterium phage FLiP OX=2023716 PE=4 SV=1
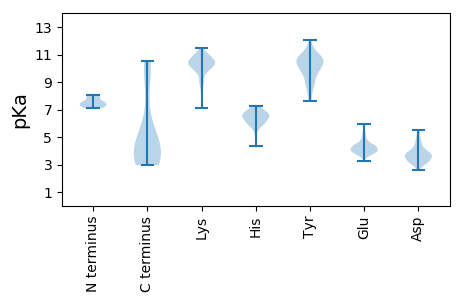
MM1 pKa = 7.5SVLAFQKK8 pKa = 10.8LSGDD12 pKa = 3.55LSTDD16 pKa = 2.96GKK18 pKa = 10.81ILVEE22 pKa = 3.95LQKK25 pKa = 11.09LNQKK29 pKa = 10.25NGLPDD34 pKa = 3.8LYY36 pKa = 11.37YY37 pKa = 10.82FFDD40 pKa = 3.16KK41 pKa = 10.73LYY43 pKa = 10.11PNRR46 pKa = 11.84EE47 pKa = 3.66PFLIEE52 pKa = 3.65RR53 pKa = 11.84SFNSGEE59 pKa = 3.75PDD61 pKa = 2.9GWFPRR66 pKa = 11.84YY67 pKa = 8.77WNNDD71 pKa = 3.45PNDD74 pKa = 3.66EE75 pKa = 4.37KK76 pKa = 11.18PFGGSFWGYY85 pKa = 8.77WNLTAFSGGEE95 pKa = 3.81NEE97 pKa = 5.11DD98 pKa = 3.66RR99 pKa = 11.84DD100 pKa = 4.23DD101 pKa = 4.26ASLVNSDD108 pKa = 3.94GQLVCYY114 pKa = 10.13VSNRR118 pKa = 11.84EE119 pKa = 3.82SHH121 pKa = 5.86ASYY124 pKa = 9.74MGEE127 pKa = 3.67ATKK130 pKa = 9.99MKK132 pKa = 10.41RR133 pKa = 11.84HH134 pKa = 5.79GGYY137 pKa = 10.06INVNAHH143 pKa = 6.02FLGFAVIDD151 pKa = 3.37QFAYY155 pKa = 8.09NTKK158 pKa = 9.93GFNGNVQIEE167 pKa = 4.39YY168 pKa = 10.71DD169 pKa = 3.85EE170 pKa = 5.08SPLEE174 pKa = 4.86KK175 pKa = 10.69GDD177 pKa = 3.46ISNGKK182 pKa = 10.1SYY184 pKa = 11.52VDD186 pKa = 3.26WFEE189 pKa = 5.96DD190 pKa = 2.88SDD192 pKa = 4.68LVVKK196 pKa = 10.55SPYY199 pKa = 8.93FFKK202 pKa = 11.05VSGEE206 pKa = 4.29GEE208 pKa = 3.98PPRR211 pKa = 11.84YY212 pKa = 8.71PEE214 pKa = 4.38KK215 pKa = 11.2VGDD218 pKa = 4.41LYY220 pKa = 11.29QFSVTADD227 pKa = 2.57IGQYY231 pKa = 10.37YY232 pKa = 9.96LIEE235 pKa = 4.43VADD238 pKa = 4.52FPYY241 pKa = 10.63FDD243 pKa = 4.54ILVDD247 pKa = 4.16FPFNAEE253 pKa = 3.82VQYY256 pKa = 11.09SYY258 pKa = 11.2AYY260 pKa = 8.58EE261 pKa = 4.06PPFIDD266 pKa = 5.37FNTVQNKK273 pKa = 8.93KK274 pKa = 10.55ASVQITQLTGFKK286 pKa = 10.25VRR288 pKa = 11.84SLTRR292 pKa = 11.84FDD294 pKa = 5.44AVDD297 pKa = 3.63SNDD300 pKa = 3.32NNTYY304 pKa = 10.1SAFVEE309 pKa = 4.35STGFYY314 pKa = 10.61EE315 pKa = 4.62MNFGVQAHH323 pKa = 6.88LDD325 pKa = 3.28KK326 pKa = 11.48SFILIRR332 pKa = 11.84NHH334 pKa = 7.17AEE336 pKa = 3.53
MM1 pKa = 7.5SVLAFQKK8 pKa = 10.8LSGDD12 pKa = 3.55LSTDD16 pKa = 2.96GKK18 pKa = 10.81ILVEE22 pKa = 3.95LQKK25 pKa = 11.09LNQKK29 pKa = 10.25NGLPDD34 pKa = 3.8LYY36 pKa = 11.37YY37 pKa = 10.82FFDD40 pKa = 3.16KK41 pKa = 10.73LYY43 pKa = 10.11PNRR46 pKa = 11.84EE47 pKa = 3.66PFLIEE52 pKa = 3.65RR53 pKa = 11.84SFNSGEE59 pKa = 3.75PDD61 pKa = 2.9GWFPRR66 pKa = 11.84YY67 pKa = 8.77WNNDD71 pKa = 3.45PNDD74 pKa = 3.66EE75 pKa = 4.37KK76 pKa = 11.18PFGGSFWGYY85 pKa = 8.77WNLTAFSGGEE95 pKa = 3.81NEE97 pKa = 5.11DD98 pKa = 3.66RR99 pKa = 11.84DD100 pKa = 4.23DD101 pKa = 4.26ASLVNSDD108 pKa = 3.94GQLVCYY114 pKa = 10.13VSNRR118 pKa = 11.84EE119 pKa = 3.82SHH121 pKa = 5.86ASYY124 pKa = 9.74MGEE127 pKa = 3.67ATKK130 pKa = 9.99MKK132 pKa = 10.41RR133 pKa = 11.84HH134 pKa = 5.79GGYY137 pKa = 10.06INVNAHH143 pKa = 6.02FLGFAVIDD151 pKa = 3.37QFAYY155 pKa = 8.09NTKK158 pKa = 9.93GFNGNVQIEE167 pKa = 4.39YY168 pKa = 10.71DD169 pKa = 3.85EE170 pKa = 5.08SPLEE174 pKa = 4.86KK175 pKa = 10.69GDD177 pKa = 3.46ISNGKK182 pKa = 10.1SYY184 pKa = 11.52VDD186 pKa = 3.26WFEE189 pKa = 5.96DD190 pKa = 2.88SDD192 pKa = 4.68LVVKK196 pKa = 10.55SPYY199 pKa = 8.93FFKK202 pKa = 11.05VSGEE206 pKa = 4.29GEE208 pKa = 3.98PPRR211 pKa = 11.84YY212 pKa = 8.71PEE214 pKa = 4.38KK215 pKa = 11.2VGDD218 pKa = 4.41LYY220 pKa = 11.29QFSVTADD227 pKa = 2.57IGQYY231 pKa = 10.37YY232 pKa = 9.96LIEE235 pKa = 4.43VADD238 pKa = 4.52FPYY241 pKa = 10.63FDD243 pKa = 4.54ILVDD247 pKa = 4.16FPFNAEE253 pKa = 3.82VQYY256 pKa = 11.09SYY258 pKa = 11.2AYY260 pKa = 8.58EE261 pKa = 4.06PPFIDD266 pKa = 5.37FNTVQNKK273 pKa = 8.93KK274 pKa = 10.55ASVQITQLTGFKK286 pKa = 10.25VRR288 pKa = 11.84SLTRR292 pKa = 11.84FDD294 pKa = 5.44AVDD297 pKa = 3.63SNDD300 pKa = 3.32NNTYY304 pKa = 10.1SAFVEE309 pKa = 4.35STGFYY314 pKa = 10.61EE315 pKa = 4.62MNFGVQAHH323 pKa = 6.88LDD325 pKa = 3.28KK326 pKa = 11.48SFILIRR332 pKa = 11.84NHH334 pKa = 7.17AEE336 pKa = 3.53
Molecular weight: 38.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A222NP96|A0A222NP96_9VIRU Uncharacterized protein OS=Flavobacterium phage FLiP OX=2023716 PE=4 SV=1
MM1 pKa = 7.29AQRR4 pKa = 11.84PRR6 pKa = 11.84SKK8 pKa = 10.46RR9 pKa = 11.84FTGFRR14 pKa = 11.84RR15 pKa = 11.84FRR17 pKa = 11.84PSFGRR22 pKa = 11.84MRR24 pKa = 11.84RR25 pKa = 11.84TYY27 pKa = 10.24SNYY30 pKa = 7.99RR31 pKa = 11.84QRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84TRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84TTTNRR44 pKa = 11.84RR45 pKa = 11.84NNMSKK50 pKa = 10.19YY51 pKa = 10.21IKK53 pKa = 9.97FAVIGVIIFLIIKK66 pKa = 9.96NKK68 pKa = 9.19TKK70 pKa = 10.64VLEE73 pKa = 4.28FFKK76 pKa = 11.09KK77 pKa = 9.55LTGKK81 pKa = 8.23KK82 pKa = 7.1TAA84 pKa = 4.13
MM1 pKa = 7.29AQRR4 pKa = 11.84PRR6 pKa = 11.84SKK8 pKa = 10.46RR9 pKa = 11.84FTGFRR14 pKa = 11.84RR15 pKa = 11.84FRR17 pKa = 11.84PSFGRR22 pKa = 11.84MRR24 pKa = 11.84RR25 pKa = 11.84TYY27 pKa = 10.24SNYY30 pKa = 7.99RR31 pKa = 11.84QRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84TRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84TTTNRR44 pKa = 11.84RR45 pKa = 11.84NNMSKK50 pKa = 10.19YY51 pKa = 10.21IKK53 pKa = 9.97FAVIGVIIFLIIKK66 pKa = 9.96NKK68 pKa = 9.19TKK70 pKa = 10.64VLEE73 pKa = 4.28FFKK76 pKa = 11.09KK77 pKa = 9.55LTGKK81 pKa = 8.23KK82 pKa = 7.1TAA84 pKa = 4.13
Molecular weight: 10.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2623 |
69 |
447 |
163.9 |
18.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.376 ± 0.628 | 0.801 ± 0.172 |
6.291 ± 0.514 | 5.719 ± 0.502 |
6.557 ± 0.598 | 4.804 ± 0.733 |
1.677 ± 0.434 | 6.634 ± 0.487 |
8.578 ± 0.61 | 8.73 ± 0.542 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.44 ± 0.21 | 6.214 ± 0.39 |
3.05 ± 0.301 | 3.584 ± 0.377 |
4.003 ± 0.821 | 6.977 ± 0.574 |
5.566 ± 0.422 | 7.053 ± 0.563 |
0.991 ± 0.191 | 4.956 ± 0.534 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
