
Sanxia tombus-like virus 8
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.71
Get precalculated fractions of proteins
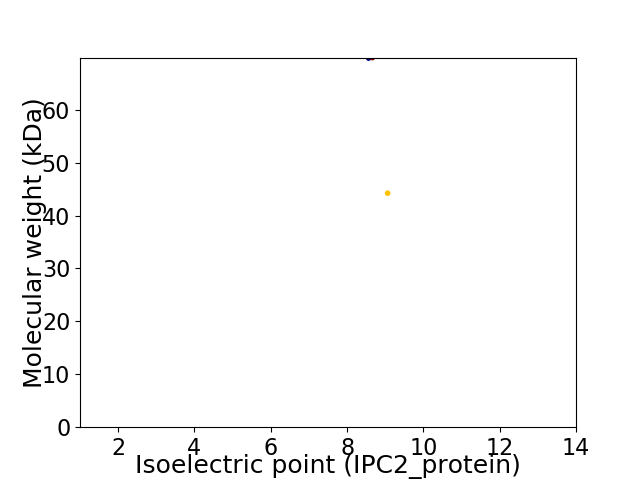
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGG5|A0A1L3KGG5_9VIRU RNA-directed RNA polymerase OS=Sanxia tombus-like virus 8 OX=1923392 PE=4 SV=1
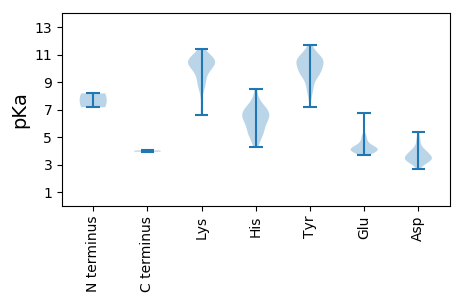
MM1 pKa = 7.16EE2 pKa = 6.71AIRR5 pKa = 11.84NIYY8 pKa = 9.56RR9 pKa = 11.84HH10 pKa = 5.38VAGAGPPPEE19 pKa = 4.36TDD21 pKa = 3.15DD22 pKa = 3.87TNLMEE27 pKa = 5.86SLEE30 pKa = 3.99QLLNANTTLNSTTIEE45 pKa = 4.18SYY47 pKa = 10.95KK48 pKa = 10.6KK49 pKa = 10.33VLGTVRR55 pKa = 11.84DD56 pKa = 3.78LTTICASLPTAATNTILDD74 pKa = 3.52EE75 pKa = 4.21RR76 pKa = 11.84LRR78 pKa = 11.84AEE80 pKa = 4.35TLTQQIAGIQHH91 pKa = 6.7GDD93 pKa = 3.37RR94 pKa = 11.84NEE96 pKa = 3.8QTAEE100 pKa = 4.09KK101 pKa = 10.42IEE103 pKa = 4.0LEE105 pKa = 3.92RR106 pKa = 11.84LKK108 pKa = 11.21AKK110 pKa = 10.47VKK112 pKa = 10.5ALEE115 pKa = 4.13EE116 pKa = 3.86QQILNDD122 pKa = 3.72RR123 pKa = 11.84ARR125 pKa = 11.84PPSIDD130 pKa = 3.35EE131 pKa = 4.21ALAFLKK137 pKa = 10.33IKK139 pKa = 10.7CLGKK143 pKa = 8.54TVDD146 pKa = 3.89EE147 pKa = 5.04TYY149 pKa = 9.8MAYY152 pKa = 10.26CGSILRR158 pKa = 11.84EE159 pKa = 3.94YY160 pKa = 10.61LLQRR164 pKa = 11.84AKK166 pKa = 10.8VSTRR170 pKa = 11.84LEE172 pKa = 3.72YY173 pKa = 10.88SIINGTMDD181 pKa = 2.93MHH183 pKa = 6.76MANIRR188 pKa = 11.84PQYY191 pKa = 10.94DD192 pKa = 3.58YY193 pKa = 11.37DD194 pKa = 3.44LHH196 pKa = 8.52NIMRR200 pKa = 11.84YY201 pKa = 8.23NDD203 pKa = 3.51CVNFVQRR210 pKa = 11.84STVRR214 pKa = 11.84VFNRR218 pKa = 11.84IYY220 pKa = 10.08TIPFTDD226 pKa = 3.22ICLKK230 pKa = 10.03SLNYY234 pKa = 9.75LRR236 pKa = 11.84PVGATEE242 pKa = 4.97NINHH246 pKa = 7.73PYY248 pKa = 9.94RR249 pKa = 11.84PRR251 pKa = 11.84FAYY254 pKa = 9.92LRR256 pKa = 11.84NILLIISILTAAYY269 pKa = 9.88NISPLFLGRR278 pKa = 11.84MKK280 pKa = 10.06HH281 pKa = 4.24TTIPNAPQPPLQPMPTGINLSTWMNIGQNILTTSSHH317 pKa = 5.76YY318 pKa = 10.67ASSSIDD324 pKa = 3.47SAHH327 pKa = 7.0GIMSSISTLWTAPASEE343 pKa = 4.63DD344 pKa = 4.23FISKK348 pKa = 10.37LSTRR352 pKa = 11.84FEE354 pKa = 3.99MDD356 pKa = 2.67EE357 pKa = 4.19KK358 pKa = 11.42SRR360 pKa = 11.84VASLLSLSLRR370 pKa = 11.84KK371 pKa = 8.98WDD373 pKa = 3.82QISTKK378 pKa = 10.65LQDD381 pKa = 3.83LSKK384 pKa = 10.73HH385 pKa = 6.3AIQHH389 pKa = 6.08SISLMDD395 pKa = 3.21ATLNHH400 pKa = 6.42SSVLTNTSYY409 pKa = 11.7NSAKK413 pKa = 10.95ALMTRR418 pKa = 11.84LEE420 pKa = 4.07PKK422 pKa = 9.04YY423 pKa = 9.51TSFLEE428 pKa = 4.25NTNTTLKK435 pKa = 9.44EE436 pKa = 4.25TIRR439 pKa = 11.84LLTHH443 pKa = 6.71TSPEE447 pKa = 4.12TCFEE451 pKa = 3.91YY452 pKa = 11.22AMNITEE458 pKa = 4.02AAINRR463 pKa = 11.84TTNFVDD469 pKa = 5.33FAAKK473 pKa = 10.47LSATRR478 pKa = 11.84LEE480 pKa = 4.12PVKK483 pKa = 11.07VKK485 pKa = 9.09ITAWSEE491 pKa = 3.88RR492 pKa = 11.84ACQEE496 pKa = 3.88TLIPAMEE503 pKa = 4.33TALSTTTYY511 pKa = 9.68SRR513 pKa = 11.84NYY515 pKa = 9.79LEE517 pKa = 4.58YY518 pKa = 10.45YY519 pKa = 9.4GLKK522 pKa = 9.19ATRR525 pKa = 11.84LLMEE529 pKa = 4.0TTLYY533 pKa = 10.88SLRR536 pKa = 11.84PVNWIVSNVNAYY548 pKa = 10.04YY549 pKa = 10.76EE550 pKa = 3.9NLIWKK555 pKa = 9.17RR556 pKa = 11.84RR557 pKa = 11.84WEE559 pKa = 3.98RR560 pKa = 11.84AQTISITLSFAAVALFITLMAIRR583 pKa = 11.84QWLSIQTVYY592 pKa = 10.59RR593 pKa = 11.84RR594 pKa = 11.84YY595 pKa = 7.87TVQPTSRR602 pKa = 11.84GRR604 pKa = 11.84TNNIPGIFRR613 pKa = 4.03
MM1 pKa = 7.16EE2 pKa = 6.71AIRR5 pKa = 11.84NIYY8 pKa = 9.56RR9 pKa = 11.84HH10 pKa = 5.38VAGAGPPPEE19 pKa = 4.36TDD21 pKa = 3.15DD22 pKa = 3.87TNLMEE27 pKa = 5.86SLEE30 pKa = 3.99QLLNANTTLNSTTIEE45 pKa = 4.18SYY47 pKa = 10.95KK48 pKa = 10.6KK49 pKa = 10.33VLGTVRR55 pKa = 11.84DD56 pKa = 3.78LTTICASLPTAATNTILDD74 pKa = 3.52EE75 pKa = 4.21RR76 pKa = 11.84LRR78 pKa = 11.84AEE80 pKa = 4.35TLTQQIAGIQHH91 pKa = 6.7GDD93 pKa = 3.37RR94 pKa = 11.84NEE96 pKa = 3.8QTAEE100 pKa = 4.09KK101 pKa = 10.42IEE103 pKa = 4.0LEE105 pKa = 3.92RR106 pKa = 11.84LKK108 pKa = 11.21AKK110 pKa = 10.47VKK112 pKa = 10.5ALEE115 pKa = 4.13EE116 pKa = 3.86QQILNDD122 pKa = 3.72RR123 pKa = 11.84ARR125 pKa = 11.84PPSIDD130 pKa = 3.35EE131 pKa = 4.21ALAFLKK137 pKa = 10.33IKK139 pKa = 10.7CLGKK143 pKa = 8.54TVDD146 pKa = 3.89EE147 pKa = 5.04TYY149 pKa = 9.8MAYY152 pKa = 10.26CGSILRR158 pKa = 11.84EE159 pKa = 3.94YY160 pKa = 10.61LLQRR164 pKa = 11.84AKK166 pKa = 10.8VSTRR170 pKa = 11.84LEE172 pKa = 3.72YY173 pKa = 10.88SIINGTMDD181 pKa = 2.93MHH183 pKa = 6.76MANIRR188 pKa = 11.84PQYY191 pKa = 10.94DD192 pKa = 3.58YY193 pKa = 11.37DD194 pKa = 3.44LHH196 pKa = 8.52NIMRR200 pKa = 11.84YY201 pKa = 8.23NDD203 pKa = 3.51CVNFVQRR210 pKa = 11.84STVRR214 pKa = 11.84VFNRR218 pKa = 11.84IYY220 pKa = 10.08TIPFTDD226 pKa = 3.22ICLKK230 pKa = 10.03SLNYY234 pKa = 9.75LRR236 pKa = 11.84PVGATEE242 pKa = 4.97NINHH246 pKa = 7.73PYY248 pKa = 9.94RR249 pKa = 11.84PRR251 pKa = 11.84FAYY254 pKa = 9.92LRR256 pKa = 11.84NILLIISILTAAYY269 pKa = 9.88NISPLFLGRR278 pKa = 11.84MKK280 pKa = 10.06HH281 pKa = 4.24TTIPNAPQPPLQPMPTGINLSTWMNIGQNILTTSSHH317 pKa = 5.76YY318 pKa = 10.67ASSSIDD324 pKa = 3.47SAHH327 pKa = 7.0GIMSSISTLWTAPASEE343 pKa = 4.63DD344 pKa = 4.23FISKK348 pKa = 10.37LSTRR352 pKa = 11.84FEE354 pKa = 3.99MDD356 pKa = 2.67EE357 pKa = 4.19KK358 pKa = 11.42SRR360 pKa = 11.84VASLLSLSLRR370 pKa = 11.84KK371 pKa = 8.98WDD373 pKa = 3.82QISTKK378 pKa = 10.65LQDD381 pKa = 3.83LSKK384 pKa = 10.73HH385 pKa = 6.3AIQHH389 pKa = 6.08SISLMDD395 pKa = 3.21ATLNHH400 pKa = 6.42SSVLTNTSYY409 pKa = 11.7NSAKK413 pKa = 10.95ALMTRR418 pKa = 11.84LEE420 pKa = 4.07PKK422 pKa = 9.04YY423 pKa = 9.51TSFLEE428 pKa = 4.25NTNTTLKK435 pKa = 9.44EE436 pKa = 4.25TIRR439 pKa = 11.84LLTHH443 pKa = 6.71TSPEE447 pKa = 4.12TCFEE451 pKa = 3.91YY452 pKa = 11.22AMNITEE458 pKa = 4.02AAINRR463 pKa = 11.84TTNFVDD469 pKa = 5.33FAAKK473 pKa = 10.47LSATRR478 pKa = 11.84LEE480 pKa = 4.12PVKK483 pKa = 11.07VKK485 pKa = 9.09ITAWSEE491 pKa = 3.88RR492 pKa = 11.84ACQEE496 pKa = 3.88TLIPAMEE503 pKa = 4.33TALSTTTYY511 pKa = 9.68SRR513 pKa = 11.84NYY515 pKa = 9.79LEE517 pKa = 4.58YY518 pKa = 10.45YY519 pKa = 9.4GLKK522 pKa = 9.19ATRR525 pKa = 11.84LLMEE529 pKa = 4.0TTLYY533 pKa = 10.88SLRR536 pKa = 11.84PVNWIVSNVNAYY548 pKa = 10.04YY549 pKa = 10.76EE550 pKa = 3.9NLIWKK555 pKa = 9.17RR556 pKa = 11.84RR557 pKa = 11.84WEE559 pKa = 3.98RR560 pKa = 11.84AQTISITLSFAAVALFITLMAIRR583 pKa = 11.84QWLSIQTVYY592 pKa = 10.59RR593 pKa = 11.84RR594 pKa = 11.84YY595 pKa = 7.87TVQPTSRR602 pKa = 11.84GRR604 pKa = 11.84TNNIPGIFRR613 pKa = 4.03
Molecular weight: 69.74 kDa
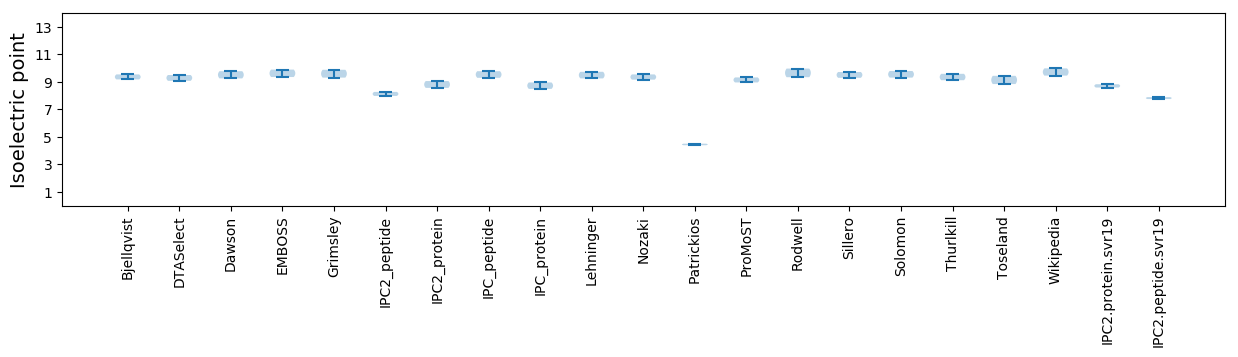
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGG5|A0A1L3KGG5_9VIRU RNA-directed RNA polymerase OS=Sanxia tombus-like virus 8 OX=1923392 PE=4 SV=1
MM1 pKa = 8.17DD2 pKa = 4.69SPSKK6 pKa = 10.51RR7 pKa = 11.84RR8 pKa = 11.84FYY10 pKa = 10.58QQVVDD15 pKa = 4.11QIRR18 pKa = 11.84NGRR21 pKa = 11.84KK22 pKa = 6.86ITSGITPFTKK32 pKa = 10.33LEE34 pKa = 4.02KK35 pKa = 10.0MGSNKK40 pKa = 9.73YY41 pKa = 9.41KK42 pKa = 10.57APRR45 pKa = 11.84LIQARR50 pKa = 11.84HH51 pKa = 4.61PTFNIAYY58 pKa = 8.4GCYY61 pKa = 9.54IKK63 pKa = 10.6PLEE66 pKa = 4.37RR67 pKa = 11.84ANKK70 pKa = 9.94HH71 pKa = 5.29KK72 pKa = 10.89LQFGKK77 pKa = 9.01GTYY80 pKa = 10.2DD81 pKa = 3.35EE82 pKa = 5.01IGAKK86 pKa = 8.45IHH88 pKa = 6.0KK89 pKa = 10.11LSRR92 pKa = 11.84KK93 pKa = 6.58YY94 pKa = 10.49QYY96 pKa = 9.28YY97 pKa = 9.31TEE99 pKa = 5.16GDD101 pKa = 3.57HH102 pKa = 6.31KK103 pKa = 9.91TFDD106 pKa = 3.29AHH108 pKa = 5.03VTRR111 pKa = 11.84DD112 pKa = 3.41MLRR115 pKa = 11.84ICHH118 pKa = 5.18EE119 pKa = 4.63HH120 pKa = 6.4YY121 pKa = 10.32RR122 pKa = 11.84SCYY125 pKa = 8.71QQDD128 pKa = 3.28NQLRR132 pKa = 11.84RR133 pKa = 11.84LCSKK137 pKa = 7.65TLRR140 pKa = 11.84NKK142 pKa = 9.85ARR144 pKa = 11.84TRR146 pKa = 11.84EE147 pKa = 4.15GEE149 pKa = 4.07NYY151 pKa = 10.1SVVGTRR157 pKa = 11.84MSGDD161 pKa = 2.98VDD163 pKa = 3.38TGYY166 pKa = 11.59GNSIINYY173 pKa = 8.77YY174 pKa = 9.58ILKK177 pKa = 9.86KK178 pKa = 10.64LLGILRR184 pKa = 11.84IKK186 pKa = 10.69GDD188 pKa = 4.27AIVNGDD194 pKa = 4.0DD195 pKa = 4.65FILFTSSKK203 pKa = 10.47LDD205 pKa = 3.36SEE207 pKa = 4.22QCKK210 pKa = 10.39RR211 pKa = 11.84ILRR214 pKa = 11.84EE215 pKa = 3.97FNMEE219 pKa = 4.03TEE221 pKa = 4.46MGEE224 pKa = 4.19STNNIHH230 pKa = 7.15HH231 pKa = 7.03IEE233 pKa = 4.14FCRR236 pKa = 11.84CRR238 pKa = 11.84LIYY241 pKa = 10.61HH242 pKa = 7.23PDD244 pKa = 3.09GHH246 pKa = 6.52PTMAFDD252 pKa = 4.94PDD254 pKa = 4.02RR255 pKa = 11.84LQKK258 pKa = 10.39IYY260 pKa = 10.73GATYY264 pKa = 10.24LPWTDD269 pKa = 3.44EE270 pKa = 3.94QYY272 pKa = 10.79TGYY275 pKa = 10.48LQVIHH280 pKa = 6.75HH281 pKa = 7.19ANFMINQNSPAHH293 pKa = 5.49RR294 pKa = 11.84QWRR297 pKa = 11.84PLLSLEE303 pKa = 4.83DD304 pKa = 3.72KK305 pKa = 10.31LKK307 pKa = 11.01KK308 pKa = 10.83YY309 pKa = 7.2MTHH312 pKa = 5.42SLEE315 pKa = 4.31RR316 pKa = 11.84VFAKK320 pKa = 10.36QSNNRR325 pKa = 11.84VYY327 pKa = 10.41KK328 pKa = 9.68WDD330 pKa = 3.91YY331 pKa = 8.44LTPSYY336 pKa = 10.87IEE338 pKa = 4.96AYY340 pKa = 9.59PNYY343 pKa = 10.28KK344 pKa = 8.72IDD346 pKa = 3.57KK347 pKa = 8.46PQYY350 pKa = 8.79PYY352 pKa = 10.52YY353 pKa = 10.77KK354 pKa = 9.87PVAPQKK360 pKa = 10.53RR361 pKa = 11.84IVINHH366 pKa = 6.48NIKK369 pKa = 10.48EE370 pKa = 4.13LFIVV374 pKa = 3.93
MM1 pKa = 8.17DD2 pKa = 4.69SPSKK6 pKa = 10.51RR7 pKa = 11.84RR8 pKa = 11.84FYY10 pKa = 10.58QQVVDD15 pKa = 4.11QIRR18 pKa = 11.84NGRR21 pKa = 11.84KK22 pKa = 6.86ITSGITPFTKK32 pKa = 10.33LEE34 pKa = 4.02KK35 pKa = 10.0MGSNKK40 pKa = 9.73YY41 pKa = 9.41KK42 pKa = 10.57APRR45 pKa = 11.84LIQARR50 pKa = 11.84HH51 pKa = 4.61PTFNIAYY58 pKa = 8.4GCYY61 pKa = 9.54IKK63 pKa = 10.6PLEE66 pKa = 4.37RR67 pKa = 11.84ANKK70 pKa = 9.94HH71 pKa = 5.29KK72 pKa = 10.89LQFGKK77 pKa = 9.01GTYY80 pKa = 10.2DD81 pKa = 3.35EE82 pKa = 5.01IGAKK86 pKa = 8.45IHH88 pKa = 6.0KK89 pKa = 10.11LSRR92 pKa = 11.84KK93 pKa = 6.58YY94 pKa = 10.49QYY96 pKa = 9.28YY97 pKa = 9.31TEE99 pKa = 5.16GDD101 pKa = 3.57HH102 pKa = 6.31KK103 pKa = 9.91TFDD106 pKa = 3.29AHH108 pKa = 5.03VTRR111 pKa = 11.84DD112 pKa = 3.41MLRR115 pKa = 11.84ICHH118 pKa = 5.18EE119 pKa = 4.63HH120 pKa = 6.4YY121 pKa = 10.32RR122 pKa = 11.84SCYY125 pKa = 8.71QQDD128 pKa = 3.28NQLRR132 pKa = 11.84RR133 pKa = 11.84LCSKK137 pKa = 7.65TLRR140 pKa = 11.84NKK142 pKa = 9.85ARR144 pKa = 11.84TRR146 pKa = 11.84EE147 pKa = 4.15GEE149 pKa = 4.07NYY151 pKa = 10.1SVVGTRR157 pKa = 11.84MSGDD161 pKa = 2.98VDD163 pKa = 3.38TGYY166 pKa = 11.59GNSIINYY173 pKa = 8.77YY174 pKa = 9.58ILKK177 pKa = 9.86KK178 pKa = 10.64LLGILRR184 pKa = 11.84IKK186 pKa = 10.69GDD188 pKa = 4.27AIVNGDD194 pKa = 4.0DD195 pKa = 4.65FILFTSSKK203 pKa = 10.47LDD205 pKa = 3.36SEE207 pKa = 4.22QCKK210 pKa = 10.39RR211 pKa = 11.84ILRR214 pKa = 11.84EE215 pKa = 3.97FNMEE219 pKa = 4.03TEE221 pKa = 4.46MGEE224 pKa = 4.19STNNIHH230 pKa = 7.15HH231 pKa = 7.03IEE233 pKa = 4.14FCRR236 pKa = 11.84CRR238 pKa = 11.84LIYY241 pKa = 10.61HH242 pKa = 7.23PDD244 pKa = 3.09GHH246 pKa = 6.52PTMAFDD252 pKa = 4.94PDD254 pKa = 4.02RR255 pKa = 11.84LQKK258 pKa = 10.39IYY260 pKa = 10.73GATYY264 pKa = 10.24LPWTDD269 pKa = 3.44EE270 pKa = 3.94QYY272 pKa = 10.79TGYY275 pKa = 10.48LQVIHH280 pKa = 6.75HH281 pKa = 7.19ANFMINQNSPAHH293 pKa = 5.49RR294 pKa = 11.84QWRR297 pKa = 11.84PLLSLEE303 pKa = 4.83DD304 pKa = 3.72KK305 pKa = 10.31LKK307 pKa = 11.01KK308 pKa = 10.83YY309 pKa = 7.2MTHH312 pKa = 5.42SLEE315 pKa = 4.31RR316 pKa = 11.84VFAKK320 pKa = 10.36QSNNRR325 pKa = 11.84VYY327 pKa = 10.41KK328 pKa = 9.68WDD330 pKa = 3.91YY331 pKa = 8.44LTPSYY336 pKa = 10.87IEE338 pKa = 4.96AYY340 pKa = 9.59PNYY343 pKa = 10.28KK344 pKa = 8.72IDD346 pKa = 3.57KK347 pKa = 8.46PQYY350 pKa = 8.79PYY352 pKa = 10.52YY353 pKa = 10.77KK354 pKa = 9.87PVAPQKK360 pKa = 10.53RR361 pKa = 11.84IVINHH366 pKa = 6.48NIKK369 pKa = 10.48EE370 pKa = 4.13LFIVV374 pKa = 3.93
Molecular weight: 44.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
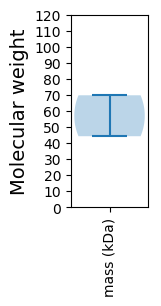
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
987 |
374 |
613 |
493.5 |
56.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.484 ± 1.498 | 1.418 ± 0.274 |
4.053 ± 0.784 | 5.37 ± 0.337 |
2.837 ± 0.387 | 3.647 ± 1.03 |
2.837 ± 0.873 | 8.004 ± 0.151 |
5.775 ± 1.684 | 9.524 ± 1.396 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.736 ± 0.199 | 5.978 ± 0.22 |
4.357 ± 0.114 | 3.951 ± 0.522 |
7.092 ± 0.239 | 6.89 ± 1.096 |
8.815 ± 1.938 | 3.546 ± 0.042 |
1.114 ± 0.189 | 5.572 ± 1.159 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
