
Halohasta litchfieldiae
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Halohasta
Average proteome isoelectric point is 4.9
Get precalculated fractions of proteins
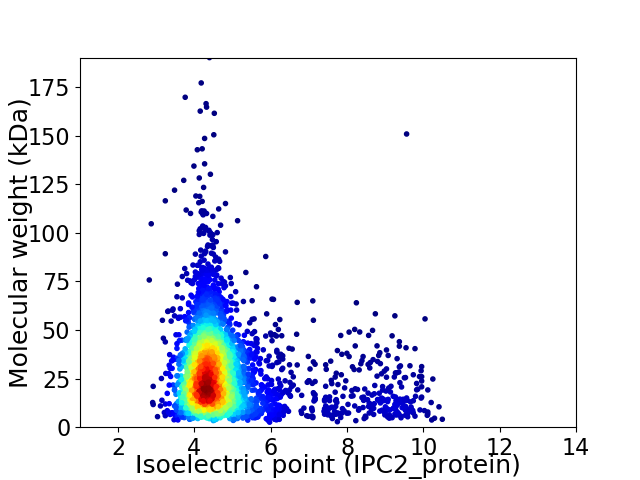
Virtual 2D-PAGE plot for 3425 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
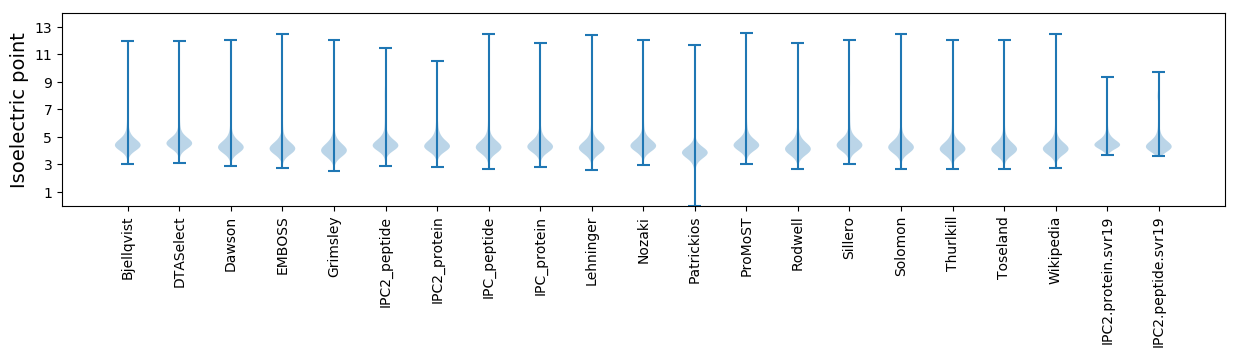
Protein with the lowest isoelectric point:
>tr|A0A1H6RCS2|A0A1H6RCS2_9EURY Adenylate cyclase OS=Halohasta litchfieldiae OX=1073996 GN=SAMN05444271_10298 PE=4 SV=1
MM1 pKa = 7.13SQDD4 pKa = 2.82ICVMVPTIRR13 pKa = 11.84EE14 pKa = 4.13YY15 pKa = 11.0EE16 pKa = 4.16CMRR19 pKa = 11.84SYY21 pKa = 10.53FQNARR26 pKa = 11.84DD27 pKa = 3.62HH28 pKa = 7.2DD29 pKa = 4.42FDD31 pKa = 4.47LSRR34 pKa = 11.84LHH36 pKa = 6.64VVLITEE42 pKa = 4.84DD43 pKa = 3.61FCDD46 pKa = 3.83TDD48 pKa = 3.52EE49 pKa = 4.3MQAMLDD55 pKa = 3.66EE56 pKa = 4.93EE57 pKa = 5.08GVDD60 pKa = 3.83GEE62 pKa = 4.54VFDD65 pKa = 4.37GSRR68 pKa = 11.84RR69 pKa = 11.84EE70 pKa = 4.1EE71 pKa = 3.86WFEE74 pKa = 3.65AHH76 pKa = 7.12DD77 pKa = 3.75VSEE80 pKa = 4.35YY81 pKa = 10.48SHH83 pKa = 6.78IVPAASHH90 pKa = 6.76AEE92 pKa = 4.07TSFGLLYY99 pKa = 9.72MWAGDD104 pKa = 3.51YY105 pKa = 10.91DD106 pKa = 4.12YY107 pKa = 11.67GFFIDD112 pKa = 6.19DD113 pKa = 3.76DD114 pKa = 4.37TLPHH118 pKa = 7.2DD119 pKa = 3.94EE120 pKa = 5.32DD121 pKa = 4.44FFGTHH126 pKa = 5.56MEE128 pKa = 3.99NLDD131 pKa = 3.65FEE133 pKa = 5.1GEE135 pKa = 4.16IEE137 pKa = 4.41SVSSDD142 pKa = 3.71EE143 pKa = 3.72QWVNVLYY150 pKa = 11.0QNAEE154 pKa = 3.85EE155 pKa = 4.46HH156 pKa = 6.72DD157 pKa = 4.72LYY159 pKa = 10.74PRR161 pKa = 11.84GYY163 pKa = 8.97PYY165 pKa = 11.08SAMHH169 pKa = 6.41EE170 pKa = 4.38SVEE173 pKa = 4.37TEE175 pKa = 3.83TTEE178 pKa = 3.54VDD180 pKa = 3.97NIVASQGLWTNVPDD194 pKa = 4.12LDD196 pKa = 4.07AVRR199 pKa = 11.84ILMDD203 pKa = 4.92GDD205 pKa = 4.0LQGQAQTRR213 pKa = 11.84TTRR216 pKa = 11.84EE217 pKa = 3.87DD218 pKa = 3.54FEE220 pKa = 6.44GDD222 pKa = 3.59FVAAEE227 pKa = 4.08DD228 pKa = 4.31NYY230 pKa = 10.86LTVCSMNLAFEE241 pKa = 4.55RR242 pKa = 11.84EE243 pKa = 4.45VIPAFYY249 pKa = 9.9QLPMDD254 pKa = 4.1EE255 pKa = 4.94NEE257 pKa = 3.9WDD259 pKa = 3.27VGRR262 pKa = 11.84FDD264 pKa = 6.36DD265 pKa = 3.27IWSGVFLKK273 pKa = 10.5RR274 pKa = 11.84ACDD277 pKa = 3.42ILDD280 pKa = 3.44KK281 pKa = 10.88QIYY284 pKa = 9.67NGLPLCEE291 pKa = 4.46HH292 pKa = 6.47NKK294 pKa = 10.1APRR297 pKa = 11.84STFDD301 pKa = 3.64DD302 pKa = 4.25LNNEE306 pKa = 4.23VPGLEE311 pKa = 4.22LNEE314 pKa = 4.33HH315 pKa = 5.94LWEE318 pKa = 4.66IVDD321 pKa = 4.41EE322 pKa = 4.31VDD324 pKa = 5.05PDD326 pKa = 4.53LSDD329 pKa = 5.26ADD331 pKa = 4.13DD332 pKa = 4.33PYY334 pKa = 12.07ARR336 pKa = 11.84IFAAMAEE343 pKa = 4.13EE344 pKa = 4.54LADD347 pKa = 4.37GDD349 pKa = 3.28WSEE352 pKa = 4.35YY353 pKa = 10.71NNGDD357 pKa = 3.32FFNYY361 pKa = 9.38IGEE364 pKa = 4.46FMGDD368 pKa = 2.91WLDD371 pKa = 3.68CLEE374 pKa = 5.21GIEE377 pKa = 4.7QAAEE381 pKa = 3.53PAANRR386 pKa = 11.84RR387 pKa = 11.84VV388 pKa = 3.2
MM1 pKa = 7.13SQDD4 pKa = 2.82ICVMVPTIRR13 pKa = 11.84EE14 pKa = 4.13YY15 pKa = 11.0EE16 pKa = 4.16CMRR19 pKa = 11.84SYY21 pKa = 10.53FQNARR26 pKa = 11.84DD27 pKa = 3.62HH28 pKa = 7.2DD29 pKa = 4.42FDD31 pKa = 4.47LSRR34 pKa = 11.84LHH36 pKa = 6.64VVLITEE42 pKa = 4.84DD43 pKa = 3.61FCDD46 pKa = 3.83TDD48 pKa = 3.52EE49 pKa = 4.3MQAMLDD55 pKa = 3.66EE56 pKa = 4.93EE57 pKa = 5.08GVDD60 pKa = 3.83GEE62 pKa = 4.54VFDD65 pKa = 4.37GSRR68 pKa = 11.84RR69 pKa = 11.84EE70 pKa = 4.1EE71 pKa = 3.86WFEE74 pKa = 3.65AHH76 pKa = 7.12DD77 pKa = 3.75VSEE80 pKa = 4.35YY81 pKa = 10.48SHH83 pKa = 6.78IVPAASHH90 pKa = 6.76AEE92 pKa = 4.07TSFGLLYY99 pKa = 9.72MWAGDD104 pKa = 3.51YY105 pKa = 10.91DD106 pKa = 4.12YY107 pKa = 11.67GFFIDD112 pKa = 6.19DD113 pKa = 3.76DD114 pKa = 4.37TLPHH118 pKa = 7.2DD119 pKa = 3.94EE120 pKa = 5.32DD121 pKa = 4.44FFGTHH126 pKa = 5.56MEE128 pKa = 3.99NLDD131 pKa = 3.65FEE133 pKa = 5.1GEE135 pKa = 4.16IEE137 pKa = 4.41SVSSDD142 pKa = 3.71EE143 pKa = 3.72QWVNVLYY150 pKa = 11.0QNAEE154 pKa = 3.85EE155 pKa = 4.46HH156 pKa = 6.72DD157 pKa = 4.72LYY159 pKa = 10.74PRR161 pKa = 11.84GYY163 pKa = 8.97PYY165 pKa = 11.08SAMHH169 pKa = 6.41EE170 pKa = 4.38SVEE173 pKa = 4.37TEE175 pKa = 3.83TTEE178 pKa = 3.54VDD180 pKa = 3.97NIVASQGLWTNVPDD194 pKa = 4.12LDD196 pKa = 4.07AVRR199 pKa = 11.84ILMDD203 pKa = 4.92GDD205 pKa = 4.0LQGQAQTRR213 pKa = 11.84TTRR216 pKa = 11.84EE217 pKa = 3.87DD218 pKa = 3.54FEE220 pKa = 6.44GDD222 pKa = 3.59FVAAEE227 pKa = 4.08DD228 pKa = 4.31NYY230 pKa = 10.86LTVCSMNLAFEE241 pKa = 4.55RR242 pKa = 11.84EE243 pKa = 4.45VIPAFYY249 pKa = 9.9QLPMDD254 pKa = 4.1EE255 pKa = 4.94NEE257 pKa = 3.9WDD259 pKa = 3.27VGRR262 pKa = 11.84FDD264 pKa = 6.36DD265 pKa = 3.27IWSGVFLKK273 pKa = 10.5RR274 pKa = 11.84ACDD277 pKa = 3.42ILDD280 pKa = 3.44KK281 pKa = 10.88QIYY284 pKa = 9.67NGLPLCEE291 pKa = 4.46HH292 pKa = 6.47NKK294 pKa = 10.1APRR297 pKa = 11.84STFDD301 pKa = 3.64DD302 pKa = 4.25LNNEE306 pKa = 4.23VPGLEE311 pKa = 4.22LNEE314 pKa = 4.33HH315 pKa = 5.94LWEE318 pKa = 4.66IVDD321 pKa = 4.41EE322 pKa = 4.31VDD324 pKa = 5.05PDD326 pKa = 4.53LSDD329 pKa = 5.26ADD331 pKa = 4.13DD332 pKa = 4.33PYY334 pKa = 12.07ARR336 pKa = 11.84IFAAMAEE343 pKa = 4.13EE344 pKa = 4.54LADD347 pKa = 4.37GDD349 pKa = 3.28WSEE352 pKa = 4.35YY353 pKa = 10.71NNGDD357 pKa = 3.32FFNYY361 pKa = 9.38IGEE364 pKa = 4.46FMGDD368 pKa = 2.91WLDD371 pKa = 3.68CLEE374 pKa = 5.21GIEE377 pKa = 4.7QAAEE381 pKa = 3.53PAANRR386 pKa = 11.84RR387 pKa = 11.84VV388 pKa = 3.2
Molecular weight: 44.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H6TYJ6|A0A1H6TYJ6_9EURY Xaa-Pro aminopeptidase OS=Halohasta litchfieldiae OX=1073996 GN=SAMN05444271_10959 PE=4 SV=1
MM1 pKa = 6.94THH3 pKa = 6.1TAIKK7 pKa = 9.46RR8 pKa = 11.84QKK10 pKa = 10.09FVIRR14 pKa = 11.84HH15 pKa = 4.72LRR17 pKa = 11.84SVNSLYY23 pKa = 10.93AQLVDD28 pKa = 3.42AVVGFGCEE36 pKa = 3.63VDD38 pKa = 3.68GVLQIRR44 pKa = 11.84PKK46 pKa = 10.49RR47 pKa = 3.66
MM1 pKa = 6.94THH3 pKa = 6.1TAIKK7 pKa = 9.46RR8 pKa = 11.84QKK10 pKa = 10.09FVIRR14 pKa = 11.84HH15 pKa = 4.72LRR17 pKa = 11.84SVNSLYY23 pKa = 10.93AQLVDD28 pKa = 3.42AVVGFGCEE36 pKa = 3.63VDD38 pKa = 3.68GVLQIRR44 pKa = 11.84PKK46 pKa = 10.49RR47 pKa = 3.66
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
947826 |
24 |
1710 |
276.7 |
30.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.719 ± 0.045 | 0.76 ± 0.015 |
8.214 ± 0.05 | 8.438 ± 0.055 |
3.309 ± 0.031 | 7.975 ± 0.042 |
1.997 ± 0.021 | 5.037 ± 0.035 |
2.251 ± 0.032 | 8.943 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.786 ± 0.018 | 2.694 ± 0.028 |
4.488 ± 0.03 | 3.157 ± 0.028 |
5.915 ± 0.041 | 6.259 ± 0.036 |
6.901 ± 0.039 | 8.367 ± 0.04 |
1.093 ± 0.016 | 2.696 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
