
Strawberry polerovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; unclassified Polerovirus
Average proteome isoelectric point is 7.46
Get precalculated fractions of proteins
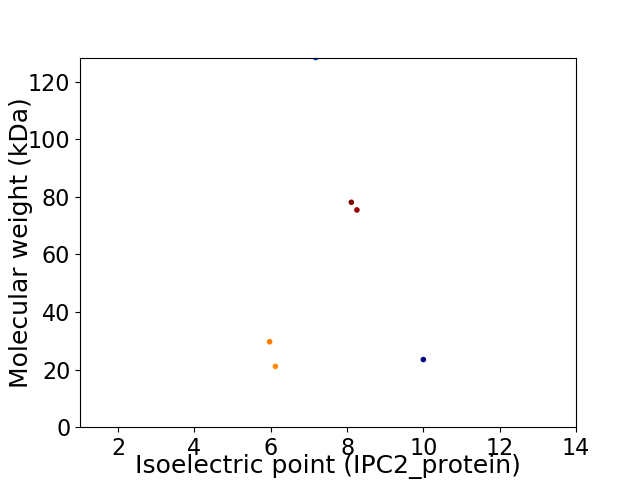
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A097RN44|A0A097RN44_9LUTE Readthrough protein OS=Strawberry polerovirus 1 OX=1564903 PE=3 SV=1
MM1 pKa = 7.38FEE3 pKa = 4.27VKK5 pKa = 10.45FGYY8 pKa = 9.97LVPTLFEE15 pKa = 4.16VTDD18 pKa = 3.55LHH20 pKa = 7.23DD21 pKa = 3.99FADD24 pKa = 3.93AFDD27 pKa = 4.13AFEE30 pKa = 4.07GLRR33 pKa = 11.84FHH35 pKa = 7.52ALLQEE40 pKa = 3.83VRR42 pKa = 11.84KK43 pKa = 9.05YY44 pKa = 11.14NRR46 pKa = 11.84VGFSRR51 pKa = 11.84SNYY54 pKa = 10.06AVDD57 pKa = 3.91RR58 pKa = 11.84DD59 pKa = 3.84VNLAFIFYY67 pKa = 10.37LPLLTQPGNKK77 pKa = 8.47PQPGDD82 pKa = 3.7SFSVPLARR90 pKa = 11.84YY91 pKa = 8.02KK92 pKa = 10.51QLIKK96 pKa = 9.76WALAVGYY103 pKa = 9.67FPEE106 pKa = 4.76LRR108 pKa = 11.84EE109 pKa = 4.41LPTTVQVCCTHH120 pKa = 7.17RR121 pKa = 11.84STEE124 pKa = 3.4TDD126 pKa = 3.08YY127 pKa = 11.25RR128 pKa = 11.84RR129 pKa = 11.84KK130 pKa = 10.13LWEE133 pKa = 4.92LDD135 pKa = 3.47TADD138 pKa = 3.28VAKK141 pKa = 10.81DD142 pKa = 3.56VVGCKK147 pKa = 9.82HH148 pKa = 6.48LPLLGANTLANIMCQRR164 pKa = 11.84FKK166 pKa = 11.25LLQDD170 pKa = 3.2EE171 pKa = 4.36HH172 pKa = 8.08RR173 pKa = 11.84KK174 pKa = 9.15IVPITRR180 pKa = 11.84NARR183 pKa = 11.84FEE185 pKa = 3.98VHH187 pKa = 6.9HH188 pKa = 6.33IHH190 pKa = 7.21LVLCLKK196 pKa = 10.85DD197 pKa = 4.49DD198 pKa = 3.71MGFLMEE204 pKa = 5.42FDD206 pKa = 4.55YY207 pKa = 11.3EE208 pKa = 4.09PHH210 pKa = 6.44CRR212 pKa = 11.84RR213 pKa = 11.84FTLDD217 pKa = 2.86VARR220 pKa = 11.84RR221 pKa = 11.84LHH223 pKa = 6.42GFGFQRR229 pKa = 11.84SAMDD233 pKa = 3.03IFEE236 pKa = 4.37TSFLRR241 pKa = 11.84SDD243 pKa = 2.86ITMEE247 pKa = 4.39DD248 pKa = 3.4LEE250 pKa = 4.56AFYY253 pKa = 10.42PP254 pKa = 3.92
MM1 pKa = 7.38FEE3 pKa = 4.27VKK5 pKa = 10.45FGYY8 pKa = 9.97LVPTLFEE15 pKa = 4.16VTDD18 pKa = 3.55LHH20 pKa = 7.23DD21 pKa = 3.99FADD24 pKa = 3.93AFDD27 pKa = 4.13AFEE30 pKa = 4.07GLRR33 pKa = 11.84FHH35 pKa = 7.52ALLQEE40 pKa = 3.83VRR42 pKa = 11.84KK43 pKa = 9.05YY44 pKa = 11.14NRR46 pKa = 11.84VGFSRR51 pKa = 11.84SNYY54 pKa = 10.06AVDD57 pKa = 3.91RR58 pKa = 11.84DD59 pKa = 3.84VNLAFIFYY67 pKa = 10.37LPLLTQPGNKK77 pKa = 8.47PQPGDD82 pKa = 3.7SFSVPLARR90 pKa = 11.84YY91 pKa = 8.02KK92 pKa = 10.51QLIKK96 pKa = 9.76WALAVGYY103 pKa = 9.67FPEE106 pKa = 4.76LRR108 pKa = 11.84EE109 pKa = 4.41LPTTVQVCCTHH120 pKa = 7.17RR121 pKa = 11.84STEE124 pKa = 3.4TDD126 pKa = 3.08YY127 pKa = 11.25RR128 pKa = 11.84RR129 pKa = 11.84KK130 pKa = 10.13LWEE133 pKa = 4.92LDD135 pKa = 3.47TADD138 pKa = 3.28VAKK141 pKa = 10.81DD142 pKa = 3.56VVGCKK147 pKa = 9.82HH148 pKa = 6.48LPLLGANTLANIMCQRR164 pKa = 11.84FKK166 pKa = 11.25LLQDD170 pKa = 3.2EE171 pKa = 4.36HH172 pKa = 8.08RR173 pKa = 11.84KK174 pKa = 9.15IVPITRR180 pKa = 11.84NARR183 pKa = 11.84FEE185 pKa = 3.98VHH187 pKa = 6.9HH188 pKa = 6.33IHH190 pKa = 7.21LVLCLKK196 pKa = 10.85DD197 pKa = 4.49DD198 pKa = 3.71MGFLMEE204 pKa = 5.42FDD206 pKa = 4.55YY207 pKa = 11.3EE208 pKa = 4.09PHH210 pKa = 6.44CRR212 pKa = 11.84RR213 pKa = 11.84FTLDD217 pKa = 2.86VARR220 pKa = 11.84RR221 pKa = 11.84LHH223 pKa = 6.42GFGFQRR229 pKa = 11.84SAMDD233 pKa = 3.03IFEE236 pKa = 4.37TSFLRR241 pKa = 11.84SDD243 pKa = 2.86ITMEE247 pKa = 4.39DD248 pKa = 3.4LEE250 pKa = 4.56AFYY253 pKa = 10.42PP254 pKa = 3.92
Molecular weight: 29.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A097RN38|A0A097RN38_9LUTE p0 OS=Strawberry polerovirus 1 OX=1564903 PE=4 SV=1
MM1 pKa = 7.54ARR3 pKa = 11.84RR4 pKa = 11.84PRR6 pKa = 11.84GSRR9 pKa = 11.84GGKK12 pKa = 9.11GKK14 pKa = 10.44KK15 pKa = 8.92KK16 pKa = 10.24QNAGRR21 pKa = 11.84RR22 pKa = 11.84NPSRR26 pKa = 11.84MARR29 pKa = 11.84PAQPIYY35 pKa = 10.63VVQRR39 pKa = 11.84PPNQGTGRR47 pKa = 11.84RR48 pKa = 11.84GRR50 pKa = 11.84RR51 pKa = 11.84NRR53 pKa = 11.84NRR55 pKa = 11.84RR56 pKa = 11.84LRR58 pKa = 11.84AQGYY62 pKa = 8.54GLGGKK67 pKa = 9.33HH68 pKa = 5.34EE69 pKa = 4.31TLKK72 pKa = 9.85FTKK75 pKa = 10.25EE76 pKa = 3.82DD77 pKa = 3.04IKK79 pKa = 11.14RR80 pKa = 11.84NSSGYY85 pKa = 7.97ITFGPSLSEE94 pKa = 4.34HH95 pKa = 6.52PAFSNGNLKK104 pKa = 10.32AYY106 pKa = 9.04KK107 pKa = 9.45HH108 pKa = 5.09YY109 pKa = 10.53RR110 pKa = 11.84ITRR113 pKa = 11.84INVEE117 pKa = 4.24YY118 pKa = 10.49LSEE121 pKa = 4.3STDD124 pKa = 3.52QAPGAIKK131 pKa = 10.63FEE133 pKa = 4.39MDD135 pKa = 3.0TSLSATKK142 pKa = 9.19VTSPIHH148 pKa = 6.15RR149 pKa = 11.84FPIKK153 pKa = 10.54KK154 pKa = 9.58NGRR157 pKa = 11.84YY158 pKa = 6.96TWTAPHH164 pKa = 6.97INGQLTRR171 pKa = 11.84EE172 pKa = 4.29TTSDD176 pKa = 2.92QCRR179 pKa = 11.84FLYY182 pKa = 10.42EE183 pKa = 3.8ATTTSTEE190 pKa = 3.82PAGVFNFTYY199 pKa = 10.2DD200 pKa = 3.29VHH202 pKa = 8.64LMDD205 pKa = 5.52NKK207 pKa = 10.96
MM1 pKa = 7.54ARR3 pKa = 11.84RR4 pKa = 11.84PRR6 pKa = 11.84GSRR9 pKa = 11.84GGKK12 pKa = 9.11GKK14 pKa = 10.44KK15 pKa = 8.92KK16 pKa = 10.24QNAGRR21 pKa = 11.84RR22 pKa = 11.84NPSRR26 pKa = 11.84MARR29 pKa = 11.84PAQPIYY35 pKa = 10.63VVQRR39 pKa = 11.84PPNQGTGRR47 pKa = 11.84RR48 pKa = 11.84GRR50 pKa = 11.84RR51 pKa = 11.84NRR53 pKa = 11.84NRR55 pKa = 11.84RR56 pKa = 11.84LRR58 pKa = 11.84AQGYY62 pKa = 8.54GLGGKK67 pKa = 9.33HH68 pKa = 5.34EE69 pKa = 4.31TLKK72 pKa = 9.85FTKK75 pKa = 10.25EE76 pKa = 3.82DD77 pKa = 3.04IKK79 pKa = 11.14RR80 pKa = 11.84NSSGYY85 pKa = 7.97ITFGPSLSEE94 pKa = 4.34HH95 pKa = 6.52PAFSNGNLKK104 pKa = 10.32AYY106 pKa = 9.04KK107 pKa = 9.45HH108 pKa = 5.09YY109 pKa = 10.53RR110 pKa = 11.84ITRR113 pKa = 11.84INVEE117 pKa = 4.24YY118 pKa = 10.49LSEE121 pKa = 4.3STDD124 pKa = 3.52QAPGAIKK131 pKa = 10.63FEE133 pKa = 4.39MDD135 pKa = 3.0TSLSATKK142 pKa = 9.19VTSPIHH148 pKa = 6.15RR149 pKa = 11.84FPIKK153 pKa = 10.54KK154 pKa = 9.58NGRR157 pKa = 11.84YY158 pKa = 6.96TWTAPHH164 pKa = 6.97INGQLTRR171 pKa = 11.84EE172 pKa = 4.29TTSDD176 pKa = 2.92QCRR179 pKa = 11.84FLYY182 pKa = 10.42EE183 pKa = 3.8ATTTSTEE190 pKa = 3.82PAGVFNFTYY199 pKa = 10.2DD200 pKa = 3.29VHH202 pKa = 8.64LMDD205 pKa = 5.52NKK207 pKa = 10.96
Molecular weight: 23.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3155 |
185 |
1139 |
525.8 |
59.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.498 ± 0.39 | 1.585 ± 0.32 |
4.913 ± 0.435 | 6.022 ± 0.299 |
3.74 ± 0.534 | 5.769 ± 0.368 |
2.536 ± 0.188 | 4.976 ± 0.443 |
5.547 ± 0.31 | 8.336 ± 1.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.87 ± 0.178 | 4.216 ± 0.33 |
6.973 ± 0.715 | 3.518 ± 0.459 |
6.815 ± 0.857 | 9.002 ± 0.901 |
6.656 ± 0.596 | 5.42 ± 0.608 |
1.902 ± 0.228 | 3.708 ± 0.195 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
