
Microbacterium lemovicicum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
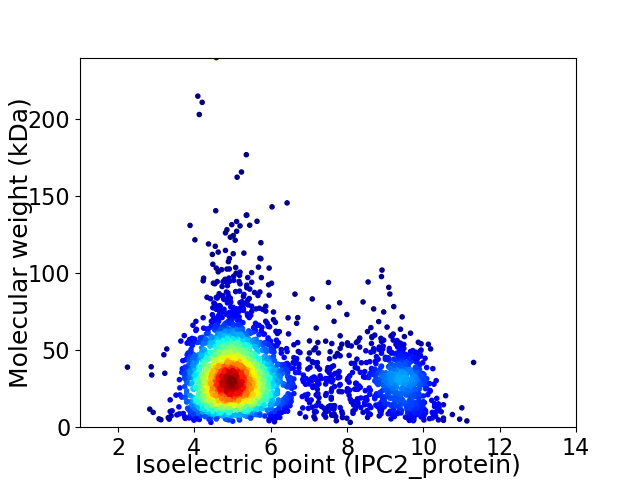
Virtual 2D-PAGE plot for 3276 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S9W656|A0A3S9W656_9MICO Uncharacterized protein OS=Microbacterium lemovicicum OX=1072463 GN=CVS47_00162 PE=4 SV=1
MM1 pKa = 6.93NTRR4 pKa = 11.84HH5 pKa = 5.38LTGTVALGAVAALALAGCASGNAAAGGSGDD35 pKa = 4.54AGDD38 pKa = 4.78KK39 pKa = 10.18GTITLGFLPSWTDD52 pKa = 3.64GLSTAYY58 pKa = 10.72LLEE61 pKa = 4.16NRR63 pKa = 11.84LEE65 pKa = 4.29GLGYY69 pKa = 8.06TVEE72 pKa = 4.2MQTLTEE78 pKa = 4.67AGPLYY83 pKa = 10.51TGLAQGDD90 pKa = 3.41IDD92 pKa = 4.7IYY94 pKa = 10.3PSAWPEE100 pKa = 3.99LTHH103 pKa = 6.41ASYY106 pKa = 9.96MEE108 pKa = 4.34KK109 pKa = 10.65YY110 pKa = 10.09GDD112 pKa = 4.02SIEE115 pKa = 5.29DD116 pKa = 3.19LGAYY120 pKa = 10.09YY121 pKa = 10.7DD122 pKa = 4.07DD123 pKa = 4.75AKK125 pKa = 10.67LTIAVPDD132 pKa = 3.81YY133 pKa = 11.63VDD135 pKa = 4.11IDD137 pKa = 5.14SIDD140 pKa = 3.63QLAGQADD147 pKa = 4.3RR148 pKa = 11.84FGGRR152 pKa = 11.84IIGIEE157 pKa = 3.98PGAGLTKK164 pKa = 9.39QTQEE168 pKa = 4.12SMLPGYY174 pKa = 10.41GLDD177 pKa = 3.79GEE179 pKa = 4.81YY180 pKa = 11.05EE181 pKa = 4.4LVTSSTAAMLTEE193 pKa = 4.92LDD195 pKa = 3.93NAITAQKK202 pKa = 10.4DD203 pKa = 3.18IAVTLWRR210 pKa = 11.84PFWANDD216 pKa = 3.3AYY218 pKa = 10.63DD219 pKa = 4.13VKK221 pKa = 11.22DD222 pKa = 3.61LADD225 pKa = 4.27PDD227 pKa = 3.69GRR229 pKa = 11.84MGEE232 pKa = 4.17PEE234 pKa = 4.06ALHH237 pKa = 5.69FLGRR241 pKa = 11.84EE242 pKa = 4.26GLGEE246 pKa = 4.46EE247 pKa = 4.53YY248 pKa = 10.26PDD250 pKa = 3.4IAEE253 pKa = 4.92LIAGIRR259 pKa = 11.84LDD261 pKa = 3.85DD262 pKa = 3.92EE263 pKa = 4.63QYY265 pKa = 11.42GALEE269 pKa = 4.09DD270 pKa = 4.39LVVNEE275 pKa = 4.11YY276 pKa = 11.42GEE278 pKa = 4.31GRR280 pKa = 11.84EE281 pKa = 4.24AEE283 pKa = 4.86AVQAWLEE290 pKa = 3.98QYY292 pKa = 10.8PDD294 pKa = 3.76VISS297 pKa = 3.61
MM1 pKa = 6.93NTRR4 pKa = 11.84HH5 pKa = 5.38LTGTVALGAVAALALAGCASGNAAAGGSGDD35 pKa = 4.54AGDD38 pKa = 4.78KK39 pKa = 10.18GTITLGFLPSWTDD52 pKa = 3.64GLSTAYY58 pKa = 10.72LLEE61 pKa = 4.16NRR63 pKa = 11.84LEE65 pKa = 4.29GLGYY69 pKa = 8.06TVEE72 pKa = 4.2MQTLTEE78 pKa = 4.67AGPLYY83 pKa = 10.51TGLAQGDD90 pKa = 3.41IDD92 pKa = 4.7IYY94 pKa = 10.3PSAWPEE100 pKa = 3.99LTHH103 pKa = 6.41ASYY106 pKa = 9.96MEE108 pKa = 4.34KK109 pKa = 10.65YY110 pKa = 10.09GDD112 pKa = 4.02SIEE115 pKa = 5.29DD116 pKa = 3.19LGAYY120 pKa = 10.09YY121 pKa = 10.7DD122 pKa = 4.07DD123 pKa = 4.75AKK125 pKa = 10.67LTIAVPDD132 pKa = 3.81YY133 pKa = 11.63VDD135 pKa = 4.11IDD137 pKa = 5.14SIDD140 pKa = 3.63QLAGQADD147 pKa = 4.3RR148 pKa = 11.84FGGRR152 pKa = 11.84IIGIEE157 pKa = 3.98PGAGLTKK164 pKa = 9.39QTQEE168 pKa = 4.12SMLPGYY174 pKa = 10.41GLDD177 pKa = 3.79GEE179 pKa = 4.81YY180 pKa = 11.05EE181 pKa = 4.4LVTSSTAAMLTEE193 pKa = 4.92LDD195 pKa = 3.93NAITAQKK202 pKa = 10.4DD203 pKa = 3.18IAVTLWRR210 pKa = 11.84PFWANDD216 pKa = 3.3AYY218 pKa = 10.63DD219 pKa = 4.13VKK221 pKa = 11.22DD222 pKa = 3.61LADD225 pKa = 4.27PDD227 pKa = 3.69GRR229 pKa = 11.84MGEE232 pKa = 4.17PEE234 pKa = 4.06ALHH237 pKa = 5.69FLGRR241 pKa = 11.84EE242 pKa = 4.26GLGEE246 pKa = 4.46EE247 pKa = 4.53YY248 pKa = 10.26PDD250 pKa = 3.4IAEE253 pKa = 4.92LIAGIRR259 pKa = 11.84LDD261 pKa = 3.85DD262 pKa = 3.92EE263 pKa = 4.63QYY265 pKa = 11.42GALEE269 pKa = 4.09DD270 pKa = 4.39LVVNEE275 pKa = 4.11YY276 pKa = 11.42GEE278 pKa = 4.31GRR280 pKa = 11.84EE281 pKa = 4.24AEE283 pKa = 4.86AVQAWLEE290 pKa = 3.98QYY292 pKa = 10.8PDD294 pKa = 3.76VISS297 pKa = 3.61
Molecular weight: 31.69 kDa
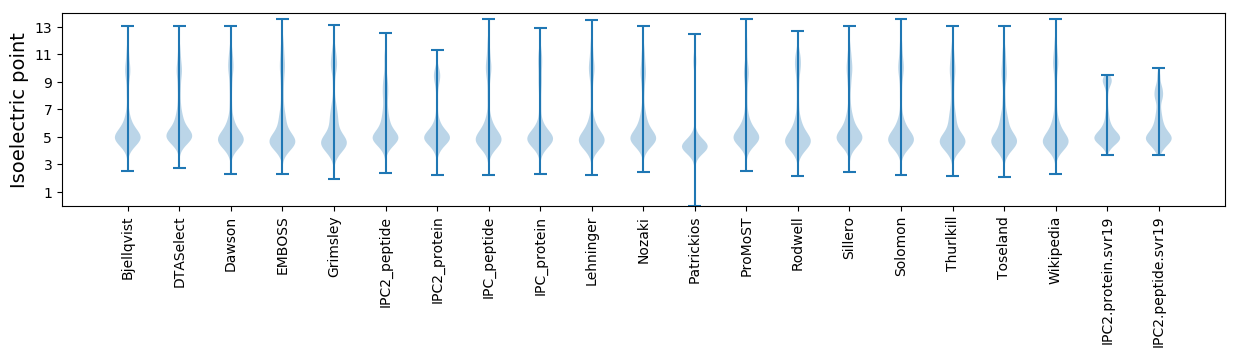
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S9W8S5|A0A3S9W8S5_9MICO ABC transporter glutamine-binding protein GlnH OS=Microbacterium lemovicicum OX=1072463 GN=glnH_3 PE=3 SV=1
MM1 pKa = 7.31VSGSRR6 pKa = 11.84TRR8 pKa = 11.84LVMASGSRR16 pKa = 11.84TRR18 pKa = 11.84LVMVSGSRR26 pKa = 11.84TRR28 pKa = 11.84LVMVSGSRR36 pKa = 11.84TRR38 pKa = 11.84LVMVSGSRR46 pKa = 11.84TRR48 pKa = 11.84LVMVSGSRR56 pKa = 11.84TRR58 pKa = 11.84LVMASGSRR66 pKa = 11.84TRR68 pKa = 11.84PVMVSASLMRR78 pKa = 11.84SATATAPMRR87 pKa = 11.84LVMASVVRR95 pKa = 11.84TRR97 pKa = 11.84RR98 pKa = 11.84AMVSGSRR105 pKa = 11.84TRR107 pKa = 11.84PVTVSGSRR115 pKa = 11.84TRR117 pKa = 11.84PVTVSGSRR125 pKa = 11.84TRR127 pKa = 11.84PVTVSGSRR135 pKa = 11.84TRR137 pKa = 11.84RR138 pKa = 11.84AMVSGSRR145 pKa = 11.84TLPVMVSGSRR155 pKa = 11.84TLPVMGTGSPTRR167 pKa = 11.84SVTATVRR174 pKa = 11.84TQLVMVSGSRR184 pKa = 11.84TRR186 pKa = 11.84PVMVSGSRR194 pKa = 11.84TRR196 pKa = 11.84PVMVSGSLTRR206 pKa = 11.84RR207 pKa = 11.84GMASVVRR214 pKa = 11.84TLPVMVSGTRR224 pKa = 11.84TRR226 pKa = 11.84PVMVSGSLTRR236 pKa = 11.84RR237 pKa = 11.84GMASVVRR244 pKa = 11.84TLPVMVSGSPTRR256 pKa = 11.84PATVSGTRR264 pKa = 11.84TRR266 pKa = 11.84PAMASVGRR274 pKa = 11.84TRR276 pKa = 11.84RR277 pKa = 11.84ARR279 pKa = 11.84GTVSPTRR286 pKa = 11.84SVTGTARR293 pKa = 11.84TQLAMASGIRR303 pKa = 11.84TRR305 pKa = 11.84RR306 pKa = 11.84GMASVVRR313 pKa = 11.84PLPVMVSGSRR323 pKa = 11.84TRR325 pKa = 11.84PVMVSGSPTLFVTVSGPRR343 pKa = 11.84TLPVTDD349 pKa = 3.48MPGRR353 pKa = 11.84TLPEE357 pKa = 3.85PVVNASTVRR366 pKa = 11.84RR367 pKa = 11.84TSFVRR372 pKa = 11.84CGLSTMTRR380 pKa = 11.84CSPTTSLPTPWTRR393 pKa = 11.84APGTSS398 pKa = 3.05
MM1 pKa = 7.31VSGSRR6 pKa = 11.84TRR8 pKa = 11.84LVMASGSRR16 pKa = 11.84TRR18 pKa = 11.84LVMVSGSRR26 pKa = 11.84TRR28 pKa = 11.84LVMVSGSRR36 pKa = 11.84TRR38 pKa = 11.84LVMVSGSRR46 pKa = 11.84TRR48 pKa = 11.84LVMVSGSRR56 pKa = 11.84TRR58 pKa = 11.84LVMASGSRR66 pKa = 11.84TRR68 pKa = 11.84PVMVSASLMRR78 pKa = 11.84SATATAPMRR87 pKa = 11.84LVMASVVRR95 pKa = 11.84TRR97 pKa = 11.84RR98 pKa = 11.84AMVSGSRR105 pKa = 11.84TRR107 pKa = 11.84PVTVSGSRR115 pKa = 11.84TRR117 pKa = 11.84PVTVSGSRR125 pKa = 11.84TRR127 pKa = 11.84PVTVSGSRR135 pKa = 11.84TRR137 pKa = 11.84RR138 pKa = 11.84AMVSGSRR145 pKa = 11.84TLPVMVSGSRR155 pKa = 11.84TLPVMGTGSPTRR167 pKa = 11.84SVTATVRR174 pKa = 11.84TQLVMVSGSRR184 pKa = 11.84TRR186 pKa = 11.84PVMVSGSRR194 pKa = 11.84TRR196 pKa = 11.84PVMVSGSLTRR206 pKa = 11.84RR207 pKa = 11.84GMASVVRR214 pKa = 11.84TLPVMVSGTRR224 pKa = 11.84TRR226 pKa = 11.84PVMVSGSLTRR236 pKa = 11.84RR237 pKa = 11.84GMASVVRR244 pKa = 11.84TLPVMVSGSPTRR256 pKa = 11.84PATVSGTRR264 pKa = 11.84TRR266 pKa = 11.84PAMASVGRR274 pKa = 11.84TRR276 pKa = 11.84RR277 pKa = 11.84ARR279 pKa = 11.84GTVSPTRR286 pKa = 11.84SVTGTARR293 pKa = 11.84TQLAMASGIRR303 pKa = 11.84TRR305 pKa = 11.84RR306 pKa = 11.84GMASVVRR313 pKa = 11.84PLPVMVSGSRR323 pKa = 11.84TRR325 pKa = 11.84PVMVSGSPTLFVTVSGPRR343 pKa = 11.84TLPVTDD349 pKa = 3.48MPGRR353 pKa = 11.84TLPEE357 pKa = 3.85PVVNASTVRR366 pKa = 11.84RR367 pKa = 11.84TSFVRR372 pKa = 11.84CGLSTMTRR380 pKa = 11.84CSPTTSLPTPWTRR393 pKa = 11.84APGTSS398 pKa = 3.05
Molecular weight: 42.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1074964 |
29 |
2343 |
328.1 |
34.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.898 ± 0.057 | 0.463 ± 0.009 |
6.542 ± 0.04 | 5.417 ± 0.04 |
3.088 ± 0.024 | 9.066 ± 0.033 |
1.905 ± 0.02 | 4.424 ± 0.032 |
1.714 ± 0.031 | 9.943 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.763 ± 0.017 | 1.773 ± 0.024 |
5.56 ± 0.03 | 2.676 ± 0.024 |
7.483 ± 0.051 | 5.658 ± 0.032 |
6.077 ± 0.038 | 9.124 ± 0.041 |
1.497 ± 0.015 | 1.929 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
