
Agromyces sp. NDB4Y10
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Agromyces; unclassified Agromyces
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
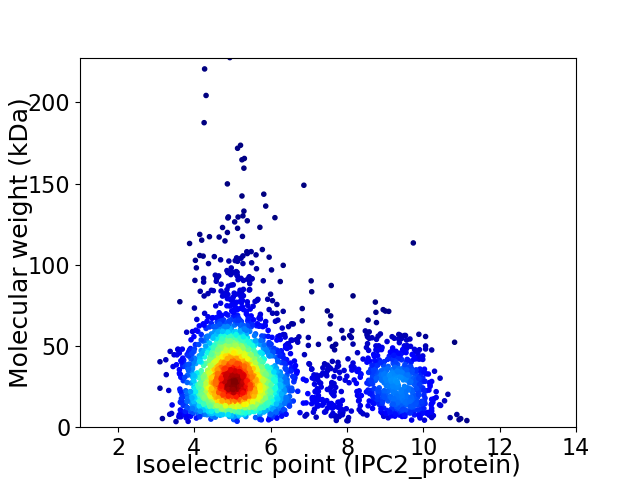
Virtual 2D-PAGE plot for 3157 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A165S8V8|A0A165S8V8_9MICO Nicotinate dehydrogenase FAD-subunit OS=Agromyces sp. NDB4Y10 OX=1775951 GN=ndhF PE=4 SV=1
MM1 pKa = 7.62GNPVLRR7 pKa = 11.84APRR10 pKa = 11.84RR11 pKa = 11.84SIGAFAVGALAASFLAATAAPANADD36 pKa = 3.38DD37 pKa = 4.81HH38 pKa = 7.12EE39 pKa = 4.63NLPGEE44 pKa = 4.2PTITLPINDD53 pKa = 3.78SAAGGTYY60 pKa = 10.54DD61 pKa = 4.24QDD63 pKa = 3.27FTRR66 pKa = 11.84AYY68 pKa = 10.6AGATPDD74 pKa = 3.42GTPVYY79 pKa = 9.93LYY81 pKa = 10.87VPAGTPLDD89 pKa = 3.73GSAPVGVDD97 pKa = 3.09SLDD100 pKa = 3.27PAFDD104 pKa = 4.52HH105 pKa = 7.13NPDD108 pKa = 4.05DD109 pKa = 5.85DD110 pKa = 4.87FVPCADD116 pKa = 4.35RR117 pKa = 11.84DD118 pKa = 4.06VADD121 pKa = 4.13FTLTQAQIDD130 pKa = 4.11YY131 pKa = 10.29MGDD134 pKa = 2.99QLANQIVAVDD144 pKa = 4.02EE145 pKa = 4.38EE146 pKa = 5.08HH147 pKa = 6.85FGPMDD152 pKa = 3.99AADD155 pKa = 4.17PDD157 pKa = 4.19VPEE160 pKa = 4.57SDD162 pKa = 3.72SLVMLVYY169 pKa = 10.51NVQDD173 pKa = 3.56DD174 pKa = 4.19NYY176 pKa = 9.32YY177 pKa = 10.9DD178 pKa = 3.98CAATSYY184 pKa = 9.15TAGYY188 pKa = 7.64FAPDD192 pKa = 5.0FIDD195 pKa = 3.48SAGMNTIVIDD205 pKa = 3.91AFDD208 pKa = 3.41WANRR212 pKa = 11.84VGPNDD217 pKa = 3.68AEE219 pKa = 4.23WRR221 pKa = 11.84DD222 pKa = 3.5GDD224 pKa = 4.35PANDD228 pKa = 4.07LPTLYY233 pKa = 10.07EE234 pKa = 3.82GVIAHH239 pKa = 6.11EE240 pKa = 4.63LEE242 pKa = 4.78HH243 pKa = 6.83LLHH246 pKa = 6.67NYY248 pKa = 9.83SDD250 pKa = 4.0PGEE253 pKa = 4.2LSWVDD258 pKa = 3.2EE259 pKa = 4.26GLADD263 pKa = 4.63FAVFLNGYY271 pKa = 9.39DD272 pKa = 3.18VGGSHH277 pKa = 6.53LTYY280 pKa = 10.21HH281 pKa = 5.09QVFYY285 pKa = 11.44AEE287 pKa = 4.24TSLTRR292 pKa = 11.84WAGGLEE298 pKa = 4.28NYY300 pKa = 8.69GAAYY304 pKa = 9.07TYY306 pKa = 8.87MQYY309 pKa = 11.01LWEE312 pKa = 4.09QAGGNGDD319 pKa = 3.68GTYY322 pKa = 10.66EE323 pKa = 4.55PDD325 pKa = 4.34LEE327 pKa = 4.13YY328 pKa = 11.16DD329 pKa = 3.67GAGGDD334 pKa = 4.39LLIKK338 pKa = 10.68LIFEE342 pKa = 4.25NQADD346 pKa = 3.78GMAGVQAAIDD356 pKa = 3.98EE357 pKa = 4.78FNAATGADD365 pKa = 3.48LRR367 pKa = 11.84SAEE370 pKa = 4.82EE371 pKa = 4.81LFQDD375 pKa = 3.53WAVAVYY381 pKa = 10.77LDD383 pKa = 5.07DD384 pKa = 4.6EE385 pKa = 4.84DD386 pKa = 5.61SEE388 pKa = 5.15RR389 pKa = 11.84FDD391 pKa = 3.49IKK393 pKa = 10.99AVDD396 pKa = 4.16FGDD399 pKa = 3.94PASTSWTIDD408 pKa = 3.03IADD411 pKa = 3.66DD412 pKa = 3.82QFWDD416 pKa = 4.14GRR418 pKa = 11.84GSNQGAQPDD427 pKa = 4.21AKK429 pKa = 10.01WEE431 pKa = 3.98RR432 pKa = 11.84RR433 pKa = 11.84KK434 pKa = 10.61NGQADD439 pKa = 3.38TALPYY444 pKa = 10.45GIQVEE449 pKa = 4.58RR450 pKa = 11.84FRR452 pKa = 11.84NPGPTVTVGLDD463 pKa = 3.46GDD465 pKa = 4.45DD466 pKa = 3.63TTQIAPHH473 pKa = 6.52TGDD476 pKa = 3.43THH478 pKa = 6.68WYY480 pKa = 9.87AGYY483 pKa = 10.35EE484 pKa = 4.15SQADD488 pKa = 3.45NVLDD492 pKa = 3.83VEE494 pKa = 4.71VADD497 pKa = 4.99GAASTLDD504 pKa = 3.23FWSWHH509 pKa = 6.06FIEE512 pKa = 5.46EE513 pKa = 4.19GWDD516 pKa = 3.44YY517 pKa = 11.6GFVEE521 pKa = 4.78ALVGDD526 pKa = 4.02EE527 pKa = 4.15WVTVPLTDD535 pKa = 4.77DD536 pKa = 3.36SGATVTTDD544 pKa = 3.24TDD546 pKa = 3.44PHH548 pKa = 6.44GNNAEE553 pKa = 4.26GNGLTGTSGGEE564 pKa = 3.98YY565 pKa = 10.19FVDD568 pKa = 3.76EE569 pKa = 4.66PEE571 pKa = 4.25YY572 pKa = 10.87VHH574 pKa = 7.26LSADD578 pKa = 3.95LPAGTTDD585 pKa = 2.73VRR587 pKa = 11.84FRR589 pKa = 11.84YY590 pKa = 8.58STDD593 pKa = 2.83AAYY596 pKa = 10.92LDD598 pKa = 4.21TGWFVDD604 pKa = 3.91DD605 pKa = 4.21VMVDD609 pKa = 3.5GEE611 pKa = 4.24AAAVSSADD619 pKa = 3.76GEE621 pKa = 4.55WIEE624 pKa = 4.49TTGLQDD630 pKa = 4.55NNWTLQVVANCDD642 pKa = 3.41LTPGVEE648 pKa = 4.21SDD650 pKa = 3.98FEE652 pKa = 4.36IVDD655 pKa = 3.38EE656 pKa = 5.01AGNHH660 pKa = 4.92VYY662 pKa = 10.43RR663 pKa = 11.84LEE665 pKa = 5.53GDD667 pKa = 4.37DD668 pKa = 4.1ISMSGLDD675 pKa = 3.51TSCAKK680 pKa = 10.46GKK682 pKa = 9.92NRR684 pKa = 11.84DD685 pKa = 3.3FATLVSNLPTGDD697 pKa = 5.35LDD699 pKa = 4.09TLDD702 pKa = 4.0AGYY705 pKa = 10.99SLGVTNTGKK714 pKa = 10.75GKK716 pKa = 10.56RR717 pKa = 11.84GG718 pKa = 3.26
MM1 pKa = 7.62GNPVLRR7 pKa = 11.84APRR10 pKa = 11.84RR11 pKa = 11.84SIGAFAVGALAASFLAATAAPANADD36 pKa = 3.38DD37 pKa = 4.81HH38 pKa = 7.12EE39 pKa = 4.63NLPGEE44 pKa = 4.2PTITLPINDD53 pKa = 3.78SAAGGTYY60 pKa = 10.54DD61 pKa = 4.24QDD63 pKa = 3.27FTRR66 pKa = 11.84AYY68 pKa = 10.6AGATPDD74 pKa = 3.42GTPVYY79 pKa = 9.93LYY81 pKa = 10.87VPAGTPLDD89 pKa = 3.73GSAPVGVDD97 pKa = 3.09SLDD100 pKa = 3.27PAFDD104 pKa = 4.52HH105 pKa = 7.13NPDD108 pKa = 4.05DD109 pKa = 5.85DD110 pKa = 4.87FVPCADD116 pKa = 4.35RR117 pKa = 11.84DD118 pKa = 4.06VADD121 pKa = 4.13FTLTQAQIDD130 pKa = 4.11YY131 pKa = 10.29MGDD134 pKa = 2.99QLANQIVAVDD144 pKa = 4.02EE145 pKa = 4.38EE146 pKa = 5.08HH147 pKa = 6.85FGPMDD152 pKa = 3.99AADD155 pKa = 4.17PDD157 pKa = 4.19VPEE160 pKa = 4.57SDD162 pKa = 3.72SLVMLVYY169 pKa = 10.51NVQDD173 pKa = 3.56DD174 pKa = 4.19NYY176 pKa = 9.32YY177 pKa = 10.9DD178 pKa = 3.98CAATSYY184 pKa = 9.15TAGYY188 pKa = 7.64FAPDD192 pKa = 5.0FIDD195 pKa = 3.48SAGMNTIVIDD205 pKa = 3.91AFDD208 pKa = 3.41WANRR212 pKa = 11.84VGPNDD217 pKa = 3.68AEE219 pKa = 4.23WRR221 pKa = 11.84DD222 pKa = 3.5GDD224 pKa = 4.35PANDD228 pKa = 4.07LPTLYY233 pKa = 10.07EE234 pKa = 3.82GVIAHH239 pKa = 6.11EE240 pKa = 4.63LEE242 pKa = 4.78HH243 pKa = 6.83LLHH246 pKa = 6.67NYY248 pKa = 9.83SDD250 pKa = 4.0PGEE253 pKa = 4.2LSWVDD258 pKa = 3.2EE259 pKa = 4.26GLADD263 pKa = 4.63FAVFLNGYY271 pKa = 9.39DD272 pKa = 3.18VGGSHH277 pKa = 6.53LTYY280 pKa = 10.21HH281 pKa = 5.09QVFYY285 pKa = 11.44AEE287 pKa = 4.24TSLTRR292 pKa = 11.84WAGGLEE298 pKa = 4.28NYY300 pKa = 8.69GAAYY304 pKa = 9.07TYY306 pKa = 8.87MQYY309 pKa = 11.01LWEE312 pKa = 4.09QAGGNGDD319 pKa = 3.68GTYY322 pKa = 10.66EE323 pKa = 4.55PDD325 pKa = 4.34LEE327 pKa = 4.13YY328 pKa = 11.16DD329 pKa = 3.67GAGGDD334 pKa = 4.39LLIKK338 pKa = 10.68LIFEE342 pKa = 4.25NQADD346 pKa = 3.78GMAGVQAAIDD356 pKa = 3.98EE357 pKa = 4.78FNAATGADD365 pKa = 3.48LRR367 pKa = 11.84SAEE370 pKa = 4.82EE371 pKa = 4.81LFQDD375 pKa = 3.53WAVAVYY381 pKa = 10.77LDD383 pKa = 5.07DD384 pKa = 4.6EE385 pKa = 4.84DD386 pKa = 5.61SEE388 pKa = 5.15RR389 pKa = 11.84FDD391 pKa = 3.49IKK393 pKa = 10.99AVDD396 pKa = 4.16FGDD399 pKa = 3.94PASTSWTIDD408 pKa = 3.03IADD411 pKa = 3.66DD412 pKa = 3.82QFWDD416 pKa = 4.14GRR418 pKa = 11.84GSNQGAQPDD427 pKa = 4.21AKK429 pKa = 10.01WEE431 pKa = 3.98RR432 pKa = 11.84RR433 pKa = 11.84KK434 pKa = 10.61NGQADD439 pKa = 3.38TALPYY444 pKa = 10.45GIQVEE449 pKa = 4.58RR450 pKa = 11.84FRR452 pKa = 11.84NPGPTVTVGLDD463 pKa = 3.46GDD465 pKa = 4.45DD466 pKa = 3.63TTQIAPHH473 pKa = 6.52TGDD476 pKa = 3.43THH478 pKa = 6.68WYY480 pKa = 9.87AGYY483 pKa = 10.35EE484 pKa = 4.15SQADD488 pKa = 3.45NVLDD492 pKa = 3.83VEE494 pKa = 4.71VADD497 pKa = 4.99GAASTLDD504 pKa = 3.23FWSWHH509 pKa = 6.06FIEE512 pKa = 5.46EE513 pKa = 4.19GWDD516 pKa = 3.44YY517 pKa = 11.6GFVEE521 pKa = 4.78ALVGDD526 pKa = 4.02EE527 pKa = 4.15WVTVPLTDD535 pKa = 4.77DD536 pKa = 3.36SGATVTTDD544 pKa = 3.24TDD546 pKa = 3.44PHH548 pKa = 6.44GNNAEE553 pKa = 4.26GNGLTGTSGGEE564 pKa = 3.98YY565 pKa = 10.19FVDD568 pKa = 3.76EE569 pKa = 4.66PEE571 pKa = 4.25YY572 pKa = 10.87VHH574 pKa = 7.26LSADD578 pKa = 3.95LPAGTTDD585 pKa = 2.73VRR587 pKa = 11.84FRR589 pKa = 11.84YY590 pKa = 8.58STDD593 pKa = 2.83AAYY596 pKa = 10.92LDD598 pKa = 4.21TGWFVDD604 pKa = 3.91DD605 pKa = 4.21VMVDD609 pKa = 3.5GEE611 pKa = 4.24AAAVSSADD619 pKa = 3.76GEE621 pKa = 4.55WIEE624 pKa = 4.49TTGLQDD630 pKa = 4.55NNWTLQVVANCDD642 pKa = 3.41LTPGVEE648 pKa = 4.21SDD650 pKa = 3.98FEE652 pKa = 4.36IVDD655 pKa = 3.38EE656 pKa = 5.01AGNHH660 pKa = 4.92VYY662 pKa = 10.43RR663 pKa = 11.84LEE665 pKa = 5.53GDD667 pKa = 4.37DD668 pKa = 4.1ISMSGLDD675 pKa = 3.51TSCAKK680 pKa = 10.46GKK682 pKa = 9.92NRR684 pKa = 11.84DD685 pKa = 3.3FATLVSNLPTGDD697 pKa = 5.35LDD699 pKa = 4.09TLDD702 pKa = 4.0AGYY705 pKa = 10.99SLGVTNTGKK714 pKa = 10.75GKK716 pKa = 10.56RR717 pKa = 11.84GG718 pKa = 3.26
Molecular weight: 77.24 kDa
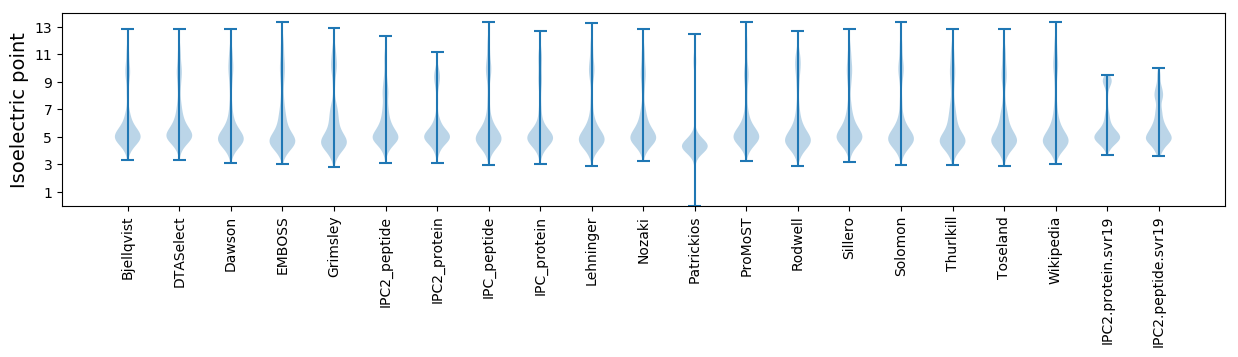
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A164DAU2|A0A164DAU2_9MICO 1 2-phenylacetyl-CoA epoxidase subunit E OS=Agromyces sp. NDB4Y10 OX=1775951 GN=paaE PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1011233 |
32 |
2103 |
320.3 |
34.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.179 ± 0.066 | 0.455 ± 0.01 |
6.454 ± 0.038 | 6.114 ± 0.038 |
3.042 ± 0.026 | 9.167 ± 0.041 |
2.023 ± 0.02 | 4.14 ± 0.031 |
1.653 ± 0.032 | 10.014 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.71 ± 0.014 | 1.744 ± 0.025 |
5.595 ± 0.033 | 2.479 ± 0.026 |
7.929 ± 0.059 | 5.056 ± 0.028 |
5.634 ± 0.035 | 9.199 ± 0.041 |
1.508 ± 0.019 | 1.904 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
