
Phreatobacter cathodiphilus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phreatobacteraceae; Phreatobacter
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
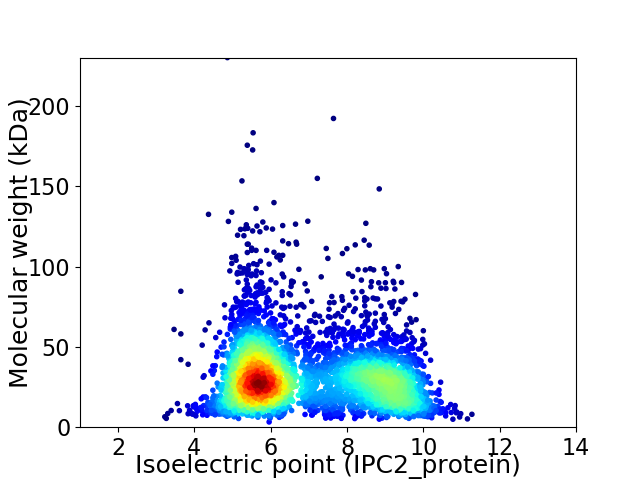
Virtual 2D-PAGE plot for 4219 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S0NBL6|A0A2S0NBL6_9PROT Asp/Glu/hydantoin racemase OS=Phreatobacter cathodiphilus OX=1868589 GN=C6569_11050 PE=4 SV=1
MM1 pKa = 7.69CNVCSGPMLRR11 pKa = 11.84DD12 pKa = 3.22WTGKK16 pKa = 7.04TVHH19 pKa = 6.71SNTGGQDD26 pKa = 3.3GPGDD30 pKa = 3.63VSFKK34 pKa = 11.04GLTGGMLQSIYY45 pKa = 11.07GGITGGVSQGVQQPTGGVSQGVTIGDD71 pKa = 3.84PTGGTAAEE79 pKa = 4.31PASGSYY85 pKa = 9.68SQSATGNTNIDD96 pKa = 4.45GILSGYY102 pKa = 9.99KK103 pKa = 7.98WTTTALTWRR112 pKa = 11.84IPTSADD118 pKa = 3.78QYY120 pKa = 10.92DD121 pKa = 4.49TDD123 pKa = 4.89PGTAGIQYY131 pKa = 10.48GDD133 pKa = 3.81TARR136 pKa = 11.84TSTFLAPTAAMTTATAEE153 pKa = 4.29ILNQMFGAVSGLSFTQAAASDD174 pKa = 3.8TTADD178 pKa = 3.51MSIGRR183 pKa = 11.84STASPSTNFNTAYY196 pKa = 10.19AYY198 pKa = 10.47YY199 pKa = 9.63PSGPGTGLSGDD210 pKa = 3.21SWYY213 pKa = 11.23SDD215 pKa = 3.23AYY217 pKa = 10.09EE218 pKa = 4.42SFGSGYY224 pKa = 10.85AYY226 pKa = 10.77SNPVKK231 pKa = 10.46GGYY234 pKa = 9.69AWTTYY239 pKa = 8.06IHH241 pKa = 7.18EE242 pKa = 4.77YY243 pKa = 8.01GHH245 pKa = 6.71NMGLKK250 pKa = 9.88HH251 pKa = 5.62GHH253 pKa = 4.39QTGGPGNTAMSADD266 pKa = 4.23RR267 pKa = 11.84DD268 pKa = 3.95SMEE271 pKa = 4.28FSVMTYY277 pKa = 9.95RR278 pKa = 11.84SYY280 pKa = 11.41VGQDD284 pKa = 2.88TVANPGYY291 pKa = 10.47QNEE294 pKa = 4.24QWGFAQSLMMYY305 pKa = 9.94DD306 pKa = 3.52IAALQVMYY314 pKa = 10.57GADD317 pKa = 3.25FTTNNTNTVYY327 pKa = 10.29TFSATTGQMFINGVGQGAPGGNRR350 pKa = 11.84VFLTIWDD357 pKa = 3.75GGGIDD362 pKa = 4.15TYY364 pKa = 11.63DD365 pKa = 3.4FSNYY369 pKa = 8.91TGNQTIDD376 pKa = 3.7LTPGSWSLMSTTQRR390 pKa = 11.84AHH392 pKa = 7.24LGQGNYY398 pKa = 10.15ARR400 pKa = 11.84GNVFNALQYY409 pKa = 10.78NGDD412 pKa = 3.52VRR414 pKa = 11.84SLIEE418 pKa = 3.82NAIGGSGNDD427 pKa = 3.81TITGNAADD435 pKa = 3.77NTLTGGAGADD445 pKa = 3.58TLIGGGGSDD454 pKa = 3.31TASYY458 pKa = 8.27ATSTSRR464 pKa = 11.84VVANLADD471 pKa = 3.81SSINTGDD478 pKa = 3.67AAGDD482 pKa = 3.74TYY484 pKa = 11.74SSIEE488 pKa = 4.03NLLGTNVSGVGDD500 pKa = 3.81ALVGNSGRR508 pKa = 11.84NFLEE512 pKa = 4.11GLGGNDD518 pKa = 3.3QLFGRR523 pKa = 11.84AGDD526 pKa = 3.72DD527 pKa = 3.36VLYY530 pKa = 11.08GGAGDD535 pKa = 4.99DD536 pKa = 4.4LLLGGLGADD545 pKa = 3.57WLDD548 pKa = 3.7GGTDD552 pKa = 3.58TDD554 pKa = 3.9TVSYY558 pKa = 10.77QEE560 pKa = 4.44AGSGVVVDD568 pKa = 4.85MLNASIQTGEE578 pKa = 3.96AAGDD582 pKa = 3.5QLISIEE588 pKa = 4.44NIFGSSFDD596 pKa = 5.35DD597 pKa = 4.05ALCGNNGRR605 pKa = 11.84NFLYY609 pKa = 10.47GQDD612 pKa = 3.84GNDD615 pKa = 3.22VLFGRR620 pKa = 11.84GGADD624 pKa = 3.1VLYY627 pKa = 10.37GGAGADD633 pKa = 3.65TLFGGAGSDD642 pKa = 3.38WLDD645 pKa = 3.31GGAGFDD651 pKa = 3.55YY652 pKa = 10.96ASYY655 pKa = 10.74LDD657 pKa = 4.38AGAAVQVDD665 pKa = 4.22MNNPAINLGDD675 pKa = 3.63AAGDD679 pKa = 3.47QLIDD683 pKa = 3.34IEE685 pKa = 4.6GLVGSGFDD693 pKa = 5.68DD694 pKa = 3.56VLLGNNLGNAIFGADD709 pKa = 4.0GNDD712 pKa = 3.38VIYY715 pKa = 10.87GRR717 pKa = 11.84GGGDD721 pKa = 3.75YY722 pKa = 10.1LTGGAGADD730 pKa = 3.15QFRR733 pKa = 11.84FANAGDD739 pKa = 4.1AGDD742 pKa = 4.63TISDD746 pKa = 4.03FQRR749 pKa = 11.84GLDD752 pKa = 3.52TLQFSASGFGGGLAGGALAANRR774 pKa = 11.84LVLGTAATQNFGQFIFDD791 pKa = 3.43SSTSILYY798 pKa = 9.44WDD800 pKa = 4.46ADD802 pKa = 3.78GTGAGAKK809 pKa = 8.98IAMMALSNINTLSTSDD825 pKa = 3.75FLINAA830 pKa = 4.81
MM1 pKa = 7.69CNVCSGPMLRR11 pKa = 11.84DD12 pKa = 3.22WTGKK16 pKa = 7.04TVHH19 pKa = 6.71SNTGGQDD26 pKa = 3.3GPGDD30 pKa = 3.63VSFKK34 pKa = 11.04GLTGGMLQSIYY45 pKa = 11.07GGITGGVSQGVQQPTGGVSQGVTIGDD71 pKa = 3.84PTGGTAAEE79 pKa = 4.31PASGSYY85 pKa = 9.68SQSATGNTNIDD96 pKa = 4.45GILSGYY102 pKa = 9.99KK103 pKa = 7.98WTTTALTWRR112 pKa = 11.84IPTSADD118 pKa = 3.78QYY120 pKa = 10.92DD121 pKa = 4.49TDD123 pKa = 4.89PGTAGIQYY131 pKa = 10.48GDD133 pKa = 3.81TARR136 pKa = 11.84TSTFLAPTAAMTTATAEE153 pKa = 4.29ILNQMFGAVSGLSFTQAAASDD174 pKa = 3.8TTADD178 pKa = 3.51MSIGRR183 pKa = 11.84STASPSTNFNTAYY196 pKa = 10.19AYY198 pKa = 10.47YY199 pKa = 9.63PSGPGTGLSGDD210 pKa = 3.21SWYY213 pKa = 11.23SDD215 pKa = 3.23AYY217 pKa = 10.09EE218 pKa = 4.42SFGSGYY224 pKa = 10.85AYY226 pKa = 10.77SNPVKK231 pKa = 10.46GGYY234 pKa = 9.69AWTTYY239 pKa = 8.06IHH241 pKa = 7.18EE242 pKa = 4.77YY243 pKa = 8.01GHH245 pKa = 6.71NMGLKK250 pKa = 9.88HH251 pKa = 5.62GHH253 pKa = 4.39QTGGPGNTAMSADD266 pKa = 4.23RR267 pKa = 11.84DD268 pKa = 3.95SMEE271 pKa = 4.28FSVMTYY277 pKa = 9.95RR278 pKa = 11.84SYY280 pKa = 11.41VGQDD284 pKa = 2.88TVANPGYY291 pKa = 10.47QNEE294 pKa = 4.24QWGFAQSLMMYY305 pKa = 9.94DD306 pKa = 3.52IAALQVMYY314 pKa = 10.57GADD317 pKa = 3.25FTTNNTNTVYY327 pKa = 10.29TFSATTGQMFINGVGQGAPGGNRR350 pKa = 11.84VFLTIWDD357 pKa = 3.75GGGIDD362 pKa = 4.15TYY364 pKa = 11.63DD365 pKa = 3.4FSNYY369 pKa = 8.91TGNQTIDD376 pKa = 3.7LTPGSWSLMSTTQRR390 pKa = 11.84AHH392 pKa = 7.24LGQGNYY398 pKa = 10.15ARR400 pKa = 11.84GNVFNALQYY409 pKa = 10.78NGDD412 pKa = 3.52VRR414 pKa = 11.84SLIEE418 pKa = 3.82NAIGGSGNDD427 pKa = 3.81TITGNAADD435 pKa = 3.77NTLTGGAGADD445 pKa = 3.58TLIGGGGSDD454 pKa = 3.31TASYY458 pKa = 8.27ATSTSRR464 pKa = 11.84VVANLADD471 pKa = 3.81SSINTGDD478 pKa = 3.67AAGDD482 pKa = 3.74TYY484 pKa = 11.74SSIEE488 pKa = 4.03NLLGTNVSGVGDD500 pKa = 3.81ALVGNSGRR508 pKa = 11.84NFLEE512 pKa = 4.11GLGGNDD518 pKa = 3.3QLFGRR523 pKa = 11.84AGDD526 pKa = 3.72DD527 pKa = 3.36VLYY530 pKa = 11.08GGAGDD535 pKa = 4.99DD536 pKa = 4.4LLLGGLGADD545 pKa = 3.57WLDD548 pKa = 3.7GGTDD552 pKa = 3.58TDD554 pKa = 3.9TVSYY558 pKa = 10.77QEE560 pKa = 4.44AGSGVVVDD568 pKa = 4.85MLNASIQTGEE578 pKa = 3.96AAGDD582 pKa = 3.5QLISIEE588 pKa = 4.44NIFGSSFDD596 pKa = 5.35DD597 pKa = 4.05ALCGNNGRR605 pKa = 11.84NFLYY609 pKa = 10.47GQDD612 pKa = 3.84GNDD615 pKa = 3.22VLFGRR620 pKa = 11.84GGADD624 pKa = 3.1VLYY627 pKa = 10.37GGAGADD633 pKa = 3.65TLFGGAGSDD642 pKa = 3.38WLDD645 pKa = 3.31GGAGFDD651 pKa = 3.55YY652 pKa = 10.96ASYY655 pKa = 10.74LDD657 pKa = 4.38AGAAVQVDD665 pKa = 4.22MNNPAINLGDD675 pKa = 3.63AAGDD679 pKa = 3.47QLIDD683 pKa = 3.34IEE685 pKa = 4.6GLVGSGFDD693 pKa = 5.68DD694 pKa = 3.56VLLGNNLGNAIFGADD709 pKa = 4.0GNDD712 pKa = 3.38VIYY715 pKa = 10.87GRR717 pKa = 11.84GGGDD721 pKa = 3.75YY722 pKa = 10.1LTGGAGADD730 pKa = 3.15QFRR733 pKa = 11.84FANAGDD739 pKa = 4.1AGDD742 pKa = 4.63TISDD746 pKa = 4.03FQRR749 pKa = 11.84GLDD752 pKa = 3.52TLQFSASGFGGGLAGGALAANRR774 pKa = 11.84LVLGTAATQNFGQFIFDD791 pKa = 3.43SSTSILYY798 pKa = 9.44WDD800 pKa = 4.46ADD802 pKa = 3.78GTGAGAKK809 pKa = 8.98IAMMALSNINTLSTSDD825 pKa = 3.75FLINAA830 pKa = 4.81
Molecular weight: 84.6 kDa
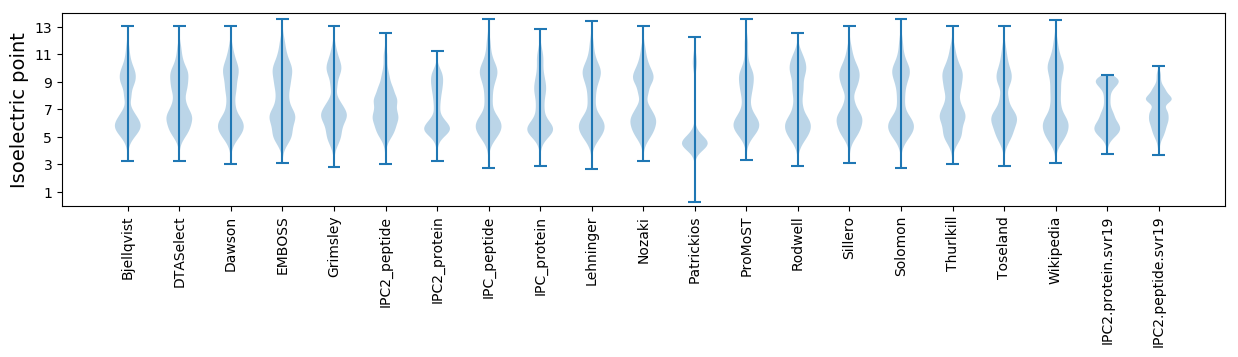
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S0NII3|A0A2S0NII3_9PROT Methyltransferase type 11 OS=Phreatobacter cathodiphilus OX=1868589 GN=C6569_20615 PE=4 SV=1
MM1 pKa = 7.65APPRR5 pKa = 11.84PRR7 pKa = 11.84RR8 pKa = 11.84WSVRR12 pKa = 11.84RR13 pKa = 11.84WPRR16 pKa = 11.84APRR19 pKa = 11.84AGPRR23 pKa = 11.84RR24 pKa = 11.84PILRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84ARR32 pKa = 11.84CRR34 pKa = 11.84ASSPRR39 pKa = 11.84SSRR42 pKa = 11.84PRR44 pKa = 11.84LSRR47 pKa = 11.84LRR49 pKa = 11.84PSPLRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84PPRR59 pKa = 11.84PTRR62 pKa = 11.84TKK64 pKa = 10.84LSAA67 pKa = 3.58
MM1 pKa = 7.65APPRR5 pKa = 11.84PRR7 pKa = 11.84RR8 pKa = 11.84WSVRR12 pKa = 11.84RR13 pKa = 11.84WPRR16 pKa = 11.84APRR19 pKa = 11.84AGPRR23 pKa = 11.84RR24 pKa = 11.84PILRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84ARR32 pKa = 11.84CRR34 pKa = 11.84ASSPRR39 pKa = 11.84SSRR42 pKa = 11.84PRR44 pKa = 11.84LSRR47 pKa = 11.84LRR49 pKa = 11.84PSPLRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84PPRR59 pKa = 11.84PTRR62 pKa = 11.84TKK64 pKa = 10.84LSAA67 pKa = 3.58
Molecular weight: 8.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1331878 |
29 |
2125 |
315.7 |
33.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.875 ± 0.06 | 0.776 ± 0.011 |
5.37 ± 0.031 | 5.373 ± 0.038 |
3.662 ± 0.026 | 9.012 ± 0.034 |
1.924 ± 0.018 | 4.868 ± 0.027 |
2.796 ± 0.031 | 9.916 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.519 ± 0.016 | 2.167 ± 0.02 |
5.615 ± 0.037 | 2.839 ± 0.025 |
7.711 ± 0.04 | 4.803 ± 0.026 |
5.449 ± 0.025 | 8.01 ± 0.027 |
1.323 ± 0.015 | 1.992 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
