
Beihai rhabdo-like virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Artoviridae; Peropuvirus; Beihai peropuvirus
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
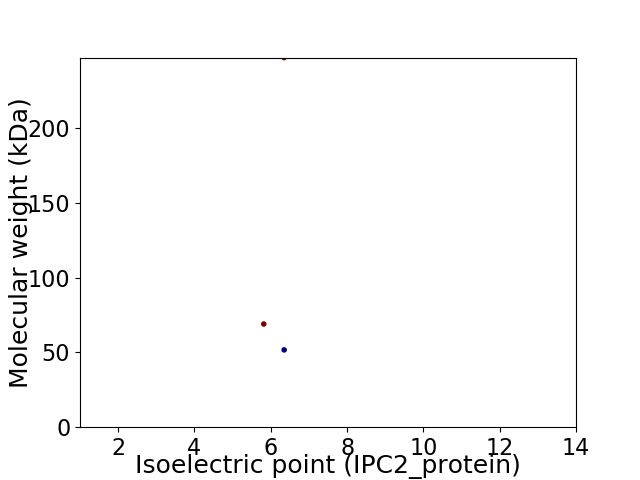
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMS9|A0A1L3KMS9_9MONO Uncharacterized protein OS=Beihai rhabdo-like virus 1 OX=1922651 PE=4 SV=1
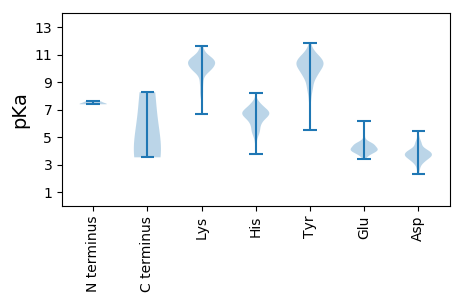
MM1 pKa = 7.4AVDD4 pKa = 3.87IGYY7 pKa = 10.51DD8 pKa = 3.45SLNKK12 pKa = 10.12VIGSHH17 pKa = 6.6PSVNYY22 pKa = 10.68AFVSGDD28 pKa = 3.08KK29 pKa = 10.93LLAKK33 pKa = 10.2KK34 pKa = 9.59ILNMTDD40 pKa = 2.52VTTIKK45 pKa = 10.57SQLEE49 pKa = 4.0APGLVVYY56 pKa = 9.84WDD58 pKa = 4.35QIPDD62 pKa = 3.56LLNAHH67 pKa = 6.32FRR69 pKa = 11.84YY70 pKa = 10.18VDD72 pKa = 3.67GLLDD76 pKa = 3.95AQDD79 pKa = 4.66FLACFYY85 pKa = 11.46AVLDD89 pKa = 3.79KK90 pKa = 11.24QYY92 pKa = 10.93FATNYY97 pKa = 9.47KK98 pKa = 10.27SVFVVSGTPAGLLVVAQHH116 pKa = 5.95FRR118 pKa = 11.84RR119 pKa = 11.84YY120 pKa = 8.43MRR122 pKa = 11.84QCDD125 pKa = 3.91IVPPDD130 pKa = 3.72ARR132 pKa = 11.84NLFLDD137 pKa = 3.96DD138 pKa = 3.7QYY140 pKa = 11.18TIPRR144 pKa = 11.84VFPASTKK151 pKa = 9.76IVPLFKK157 pKa = 10.78VEE159 pKa = 3.81PAYY162 pKa = 10.85VHH164 pKa = 7.21SFFQDD169 pKa = 3.35SNVSFDD175 pKa = 3.95SKK177 pKa = 11.01VLSSFFNNFPSTRR190 pKa = 11.84DD191 pKa = 3.27KK192 pKa = 11.13FINKK196 pKa = 6.71YY197 pKa = 8.85TIYY200 pKa = 9.94PYY202 pKa = 10.11HH203 pKa = 7.12CPASAPTTYY212 pKa = 10.08ISSPEE217 pKa = 3.93VEE219 pKa = 4.78SILMVSVASLCTAYY233 pKa = 10.36SQRR236 pKa = 11.84SRR238 pKa = 11.84ITQQIGKK245 pKa = 9.73RR246 pKa = 11.84ITAMLAGSNYY256 pKa = 9.47PSIRR260 pKa = 11.84VDD262 pKa = 4.01SINDD266 pKa = 3.44ALTAIDD272 pKa = 4.51WKK274 pKa = 10.76PEE276 pKa = 3.92EE277 pKa = 4.15QPPAIVEE284 pKa = 4.49AIGWVCAINKK294 pKa = 9.87DD295 pKa = 4.19GIIDD299 pKa = 4.0PVTSINDD306 pKa = 3.5LMDD309 pKa = 5.07PDD311 pKa = 3.58EE312 pKa = 4.21WAARR316 pKa = 11.84YY317 pKa = 7.67HH318 pKa = 6.06VPDD321 pKa = 3.04WVKK324 pKa = 10.53FLFVQMRR331 pKa = 11.84LVYY334 pKa = 10.65SHH336 pKa = 6.88FQCSSINFALVILPLIEE353 pKa = 4.86SLTTPGTSYY362 pKa = 9.85WKK364 pKa = 10.47KK365 pKa = 10.91DD366 pKa = 2.73IDD368 pKa = 3.86ALNEE372 pKa = 3.86IKK374 pKa = 10.69NIIIDD379 pKa = 4.11CPYY382 pKa = 8.72SNCMFTIPEE391 pKa = 4.03KK392 pKa = 10.56RR393 pKa = 11.84RR394 pKa = 11.84ISAFPRR400 pKa = 11.84LVYY403 pKa = 10.27MGTHH407 pKa = 5.25YY408 pKa = 10.31HH409 pKa = 6.63AKK411 pKa = 9.98HH412 pKa = 6.34LSDD415 pKa = 4.56EE416 pKa = 4.23KK417 pKa = 11.2DD418 pKa = 3.38KK419 pKa = 11.64EE420 pKa = 4.11DD421 pKa = 3.49FKK423 pKa = 11.57FYY425 pKa = 9.19VTEE428 pKa = 4.18GTVHH432 pKa = 6.47HH433 pKa = 6.58FKK435 pKa = 10.64EE436 pKa = 4.55KK437 pKa = 10.26KK438 pKa = 9.99DD439 pKa = 3.65QEE441 pKa = 4.29LCRR444 pKa = 11.84LQVQSLPRR452 pKa = 11.84PTVSALAILGSATPAEE468 pKa = 4.23KK469 pKa = 9.82MEE471 pKa = 5.06EE472 pKa = 4.09IVSGYY477 pKa = 11.26DD478 pKa = 3.34NILQAAIYY486 pKa = 10.49QEE488 pKa = 4.18MKK490 pKa = 10.38RR491 pKa = 11.84NYY493 pKa = 8.76PQAPWVTYY501 pKa = 9.92KK502 pKa = 10.56HH503 pKa = 5.86EE504 pKa = 4.35KK505 pKa = 8.77YY506 pKa = 10.3KK507 pKa = 10.32RR508 pKa = 11.84AEE510 pKa = 4.06VARR513 pKa = 11.84MRR515 pKa = 11.84TSLLNIIDD523 pKa = 3.99QKK525 pKa = 10.65YY526 pKa = 9.64QDD528 pKa = 4.67AITYY532 pKa = 10.5LNEE535 pKa = 4.44LYY537 pKa = 10.2MGAQNDD543 pKa = 3.6EE544 pKa = 3.94TRR546 pKa = 11.84IQLKK550 pKa = 8.14TKK552 pKa = 10.31KK553 pKa = 9.87DD554 pKa = 3.61DD555 pKa = 3.59FMKK558 pKa = 10.52TVLGLKK564 pKa = 10.38AKK566 pKa = 8.92LTTLKK571 pKa = 10.76EE572 pKa = 3.82EE573 pKa = 4.12MAIPEE578 pKa = 4.04NPEE581 pKa = 3.71FEE583 pKa = 4.41VEE585 pKa = 4.01RR586 pKa = 11.84EE587 pKa = 3.97KK588 pKa = 11.35LIDD591 pKa = 3.71LAKK594 pKa = 10.41PILDD598 pKa = 4.97AINAIIMM605 pKa = 4.14
MM1 pKa = 7.4AVDD4 pKa = 3.87IGYY7 pKa = 10.51DD8 pKa = 3.45SLNKK12 pKa = 10.12VIGSHH17 pKa = 6.6PSVNYY22 pKa = 10.68AFVSGDD28 pKa = 3.08KK29 pKa = 10.93LLAKK33 pKa = 10.2KK34 pKa = 9.59ILNMTDD40 pKa = 2.52VTTIKK45 pKa = 10.57SQLEE49 pKa = 4.0APGLVVYY56 pKa = 9.84WDD58 pKa = 4.35QIPDD62 pKa = 3.56LLNAHH67 pKa = 6.32FRR69 pKa = 11.84YY70 pKa = 10.18VDD72 pKa = 3.67GLLDD76 pKa = 3.95AQDD79 pKa = 4.66FLACFYY85 pKa = 11.46AVLDD89 pKa = 3.79KK90 pKa = 11.24QYY92 pKa = 10.93FATNYY97 pKa = 9.47KK98 pKa = 10.27SVFVVSGTPAGLLVVAQHH116 pKa = 5.95FRR118 pKa = 11.84RR119 pKa = 11.84YY120 pKa = 8.43MRR122 pKa = 11.84QCDD125 pKa = 3.91IVPPDD130 pKa = 3.72ARR132 pKa = 11.84NLFLDD137 pKa = 3.96DD138 pKa = 3.7QYY140 pKa = 11.18TIPRR144 pKa = 11.84VFPASTKK151 pKa = 9.76IVPLFKK157 pKa = 10.78VEE159 pKa = 3.81PAYY162 pKa = 10.85VHH164 pKa = 7.21SFFQDD169 pKa = 3.35SNVSFDD175 pKa = 3.95SKK177 pKa = 11.01VLSSFFNNFPSTRR190 pKa = 11.84DD191 pKa = 3.27KK192 pKa = 11.13FINKK196 pKa = 6.71YY197 pKa = 8.85TIYY200 pKa = 9.94PYY202 pKa = 10.11HH203 pKa = 7.12CPASAPTTYY212 pKa = 10.08ISSPEE217 pKa = 3.93VEE219 pKa = 4.78SILMVSVASLCTAYY233 pKa = 10.36SQRR236 pKa = 11.84SRR238 pKa = 11.84ITQQIGKK245 pKa = 9.73RR246 pKa = 11.84ITAMLAGSNYY256 pKa = 9.47PSIRR260 pKa = 11.84VDD262 pKa = 4.01SINDD266 pKa = 3.44ALTAIDD272 pKa = 4.51WKK274 pKa = 10.76PEE276 pKa = 3.92EE277 pKa = 4.15QPPAIVEE284 pKa = 4.49AIGWVCAINKK294 pKa = 9.87DD295 pKa = 4.19GIIDD299 pKa = 4.0PVTSINDD306 pKa = 3.5LMDD309 pKa = 5.07PDD311 pKa = 3.58EE312 pKa = 4.21WAARR316 pKa = 11.84YY317 pKa = 7.67HH318 pKa = 6.06VPDD321 pKa = 3.04WVKK324 pKa = 10.53FLFVQMRR331 pKa = 11.84LVYY334 pKa = 10.65SHH336 pKa = 6.88FQCSSINFALVILPLIEE353 pKa = 4.86SLTTPGTSYY362 pKa = 9.85WKK364 pKa = 10.47KK365 pKa = 10.91DD366 pKa = 2.73IDD368 pKa = 3.86ALNEE372 pKa = 3.86IKK374 pKa = 10.69NIIIDD379 pKa = 4.11CPYY382 pKa = 8.72SNCMFTIPEE391 pKa = 4.03KK392 pKa = 10.56RR393 pKa = 11.84RR394 pKa = 11.84ISAFPRR400 pKa = 11.84LVYY403 pKa = 10.27MGTHH407 pKa = 5.25YY408 pKa = 10.31HH409 pKa = 6.63AKK411 pKa = 9.98HH412 pKa = 6.34LSDD415 pKa = 4.56EE416 pKa = 4.23KK417 pKa = 11.2DD418 pKa = 3.38KK419 pKa = 11.64EE420 pKa = 4.11DD421 pKa = 3.49FKK423 pKa = 11.57FYY425 pKa = 9.19VTEE428 pKa = 4.18GTVHH432 pKa = 6.47HH433 pKa = 6.58FKK435 pKa = 10.64EE436 pKa = 4.55KK437 pKa = 10.26KK438 pKa = 9.99DD439 pKa = 3.65QEE441 pKa = 4.29LCRR444 pKa = 11.84LQVQSLPRR452 pKa = 11.84PTVSALAILGSATPAEE468 pKa = 4.23KK469 pKa = 9.82MEE471 pKa = 5.06EE472 pKa = 4.09IVSGYY477 pKa = 11.26DD478 pKa = 3.34NILQAAIYY486 pKa = 10.49QEE488 pKa = 4.18MKK490 pKa = 10.38RR491 pKa = 11.84NYY493 pKa = 8.76PQAPWVTYY501 pKa = 9.92KK502 pKa = 10.56HH503 pKa = 5.86EE504 pKa = 4.35KK505 pKa = 8.77YY506 pKa = 10.3KK507 pKa = 10.32RR508 pKa = 11.84AEE510 pKa = 4.06VARR513 pKa = 11.84MRR515 pKa = 11.84TSLLNIIDD523 pKa = 3.99QKK525 pKa = 10.65YY526 pKa = 9.64QDD528 pKa = 4.67AITYY532 pKa = 10.5LNEE535 pKa = 4.44LYY537 pKa = 10.2MGAQNDD543 pKa = 3.6EE544 pKa = 3.94TRR546 pKa = 11.84IQLKK550 pKa = 8.14TKK552 pKa = 10.31KK553 pKa = 9.87DD554 pKa = 3.61DD555 pKa = 3.59FMKK558 pKa = 10.52TVLGLKK564 pKa = 10.38AKK566 pKa = 8.92LTTLKK571 pKa = 10.76EE572 pKa = 3.82EE573 pKa = 4.12MAIPEE578 pKa = 4.04NPEE581 pKa = 3.71FEE583 pKa = 4.41VEE585 pKa = 4.01RR586 pKa = 11.84EE587 pKa = 3.97KK588 pKa = 11.35LIDD591 pKa = 3.71LAKK594 pKa = 10.41PILDD598 pKa = 4.97AINAIIMM605 pKa = 4.14
Molecular weight: 69.03 kDa
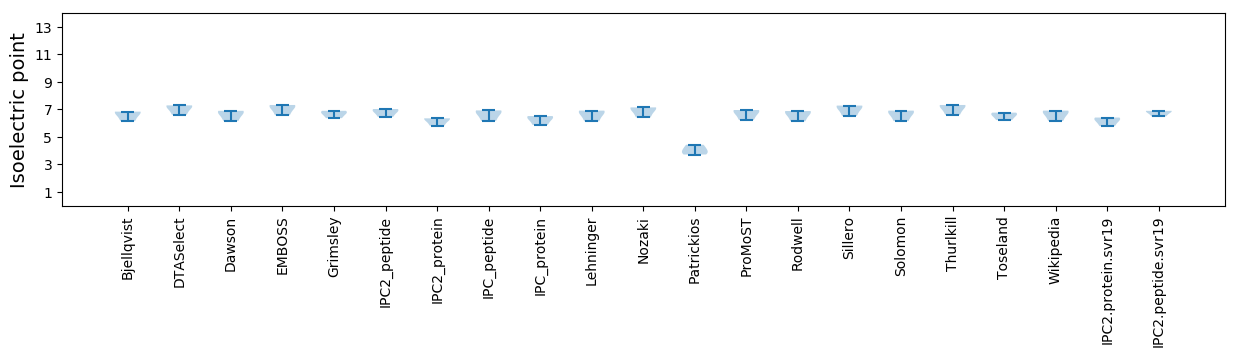
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMS9|A0A1L3KMS9_9MONO Uncharacterized protein OS=Beihai rhabdo-like virus 1 OX=1922651 PE=4 SV=1
MM1 pKa = 7.38TKK3 pKa = 10.22RR4 pKa = 11.84EE5 pKa = 4.16ILSCDD10 pKa = 3.61KK11 pKa = 10.41LRR13 pKa = 11.84EE14 pKa = 4.43MISSTSVSGPLSSQGNDD31 pKa = 2.54INMLSPLDD39 pKa = 3.71QQILNEE45 pKa = 4.02LRR47 pKa = 11.84GSEE50 pKa = 4.35SLPPSTILPVRR61 pKa = 11.84RR62 pKa = 11.84SRR64 pKa = 11.84TVEE67 pKa = 3.93DD68 pKa = 3.49LTNIPTSQPIEE79 pKa = 3.84QRR81 pKa = 11.84LVFSSPIEE89 pKa = 3.93STKK92 pKa = 10.93RR93 pKa = 11.84KK94 pKa = 7.92ITEE97 pKa = 4.47DD98 pKa = 3.64NSPMSSGTDD107 pKa = 3.22QLISKK112 pKa = 8.33LTEE115 pKa = 4.06LPLRR119 pKa = 11.84PYY121 pKa = 10.29TPLTEE126 pKa = 4.74LLPISDD132 pKa = 4.4ILDD135 pKa = 3.94PQPAATSTTRR145 pKa = 11.84SSPFKK150 pKa = 10.44KK151 pKa = 10.01RR152 pKa = 11.84STRR155 pKa = 11.84ASSVHH160 pKa = 5.16TKK162 pKa = 10.01ISYY165 pKa = 10.22VPTLPSEE172 pKa = 4.23IQEE175 pKa = 4.48SEE177 pKa = 3.83TDD179 pKa = 3.06TDD181 pKa = 3.83IEE183 pKa = 4.08VDD185 pKa = 3.8DD186 pKa = 4.23AHH188 pKa = 6.74SLSVPMSRR196 pKa = 11.84FLQASTPPPPSPSTSHH212 pKa = 6.94NLNTYY217 pKa = 9.71NLASAYY223 pKa = 9.76LAQSDD228 pKa = 4.53PIEE231 pKa = 4.18PTGNYY236 pKa = 7.35TVSSKK241 pKa = 10.01MMAFLSGEE249 pKa = 4.33SKK251 pKa = 9.83TVDD254 pKa = 2.51SDD256 pKa = 3.19MDD258 pKa = 3.96LEE260 pKa = 4.41LKK262 pKa = 10.42RR263 pKa = 11.84WATDD267 pKa = 2.63RR268 pKa = 11.84WDD270 pKa = 3.64QLINPPKK277 pKa = 10.26GLKK280 pKa = 9.9CEE282 pKa = 4.21HH283 pKa = 6.65FALCLKK289 pKa = 10.16YY290 pKa = 10.67ISGNSNRR297 pKa = 11.84HH298 pKa = 4.51KK299 pKa = 10.29AYY301 pKa = 10.41HH302 pKa = 5.46IAKK305 pKa = 9.99GNFHH309 pKa = 6.37MACATEE315 pKa = 4.43QVWSDD320 pKa = 3.6LPKK323 pKa = 9.93LTKK326 pKa = 8.36TTSNLKK332 pKa = 10.16SALSTPHH339 pKa = 5.34MTIEE343 pKa = 4.14LQSLITQHH351 pKa = 5.55TQLLQDD357 pKa = 3.82NNRR360 pKa = 11.84LITTLQEE367 pKa = 4.2YY368 pKa = 10.41GGSLINQYY376 pKa = 9.62QDD378 pKa = 2.9HH379 pKa = 6.61TNQLKK384 pKa = 10.9NITQEE389 pKa = 3.85VSNTLSSKK397 pKa = 10.22ISSLVSAIEE406 pKa = 4.04KK407 pKa = 10.7LSGLSDD413 pKa = 3.58KK414 pKa = 11.07EE415 pKa = 3.79QGKK418 pKa = 9.08LLKK421 pKa = 10.18RR422 pKa = 11.84KK423 pKa = 9.93SLGTEE428 pKa = 4.02LLDD431 pKa = 4.19SSLSSVGPSASATRR445 pKa = 11.84SQSSRR450 pKa = 11.84RR451 pKa = 11.84GSTVSTYY458 pKa = 10.22PYY460 pKa = 9.38YY461 pKa = 10.52YY462 pKa = 11.04SNVTSLPP469 pKa = 3.52
MM1 pKa = 7.38TKK3 pKa = 10.22RR4 pKa = 11.84EE5 pKa = 4.16ILSCDD10 pKa = 3.61KK11 pKa = 10.41LRR13 pKa = 11.84EE14 pKa = 4.43MISSTSVSGPLSSQGNDD31 pKa = 2.54INMLSPLDD39 pKa = 3.71QQILNEE45 pKa = 4.02LRR47 pKa = 11.84GSEE50 pKa = 4.35SLPPSTILPVRR61 pKa = 11.84RR62 pKa = 11.84SRR64 pKa = 11.84TVEE67 pKa = 3.93DD68 pKa = 3.49LTNIPTSQPIEE79 pKa = 3.84QRR81 pKa = 11.84LVFSSPIEE89 pKa = 3.93STKK92 pKa = 10.93RR93 pKa = 11.84KK94 pKa = 7.92ITEE97 pKa = 4.47DD98 pKa = 3.64NSPMSSGTDD107 pKa = 3.22QLISKK112 pKa = 8.33LTEE115 pKa = 4.06LPLRR119 pKa = 11.84PYY121 pKa = 10.29TPLTEE126 pKa = 4.74LLPISDD132 pKa = 4.4ILDD135 pKa = 3.94PQPAATSTTRR145 pKa = 11.84SSPFKK150 pKa = 10.44KK151 pKa = 10.01RR152 pKa = 11.84STRR155 pKa = 11.84ASSVHH160 pKa = 5.16TKK162 pKa = 10.01ISYY165 pKa = 10.22VPTLPSEE172 pKa = 4.23IQEE175 pKa = 4.48SEE177 pKa = 3.83TDD179 pKa = 3.06TDD181 pKa = 3.83IEE183 pKa = 4.08VDD185 pKa = 3.8DD186 pKa = 4.23AHH188 pKa = 6.74SLSVPMSRR196 pKa = 11.84FLQASTPPPPSPSTSHH212 pKa = 6.94NLNTYY217 pKa = 9.71NLASAYY223 pKa = 9.76LAQSDD228 pKa = 4.53PIEE231 pKa = 4.18PTGNYY236 pKa = 7.35TVSSKK241 pKa = 10.01MMAFLSGEE249 pKa = 4.33SKK251 pKa = 9.83TVDD254 pKa = 2.51SDD256 pKa = 3.19MDD258 pKa = 3.96LEE260 pKa = 4.41LKK262 pKa = 10.42RR263 pKa = 11.84WATDD267 pKa = 2.63RR268 pKa = 11.84WDD270 pKa = 3.64QLINPPKK277 pKa = 10.26GLKK280 pKa = 9.9CEE282 pKa = 4.21HH283 pKa = 6.65FALCLKK289 pKa = 10.16YY290 pKa = 10.67ISGNSNRR297 pKa = 11.84HH298 pKa = 4.51KK299 pKa = 10.29AYY301 pKa = 10.41HH302 pKa = 5.46IAKK305 pKa = 9.99GNFHH309 pKa = 6.37MACATEE315 pKa = 4.43QVWSDD320 pKa = 3.6LPKK323 pKa = 9.93LTKK326 pKa = 8.36TTSNLKK332 pKa = 10.16SALSTPHH339 pKa = 5.34MTIEE343 pKa = 4.14LQSLITQHH351 pKa = 5.55TQLLQDD357 pKa = 3.82NNRR360 pKa = 11.84LITTLQEE367 pKa = 4.2YY368 pKa = 10.41GGSLINQYY376 pKa = 9.62QDD378 pKa = 2.9HH379 pKa = 6.61TNQLKK384 pKa = 10.9NITQEE389 pKa = 3.85VSNTLSSKK397 pKa = 10.22ISSLVSAIEE406 pKa = 4.04KK407 pKa = 10.7LSGLSDD413 pKa = 3.58KK414 pKa = 11.07EE415 pKa = 3.79QGKK418 pKa = 9.08LLKK421 pKa = 10.18RR422 pKa = 11.84KK423 pKa = 9.93SLGTEE428 pKa = 4.02LLDD431 pKa = 4.19SSLSSVGPSASATRR445 pKa = 11.84SQSSRR450 pKa = 11.84RR451 pKa = 11.84GSTVSTYY458 pKa = 10.22PYY460 pKa = 9.38YY461 pKa = 10.52YY462 pKa = 11.04SNVTSLPP469 pKa = 3.52
Molecular weight: 51.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3247 |
469 |
2173 |
1082.3 |
122.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.928 ± 0.818 | 1.848 ± 0.382 |
5.082 ± 0.516 | 4.527 ± 0.381 |
4.558 ± 1.037 | 4.373 ± 0.642 |
3.08 ± 0.508 | 7.545 ± 0.595 |
4.958 ± 0.732 | 11.426 ± 0.966 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.033 ± 0.21 | 4.743 ± 0.244 |
4.774 ± 0.912 | 5.112 ± 0.398 |
4.158 ± 0.093 | 9.701 ± 1.744 |
6.19 ± 1.006 | 5.698 ± 0.666 |
1.17 ± 0.171 | 4.096 ± 0.404 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
