
Hondaea fermentalgiana
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Bigyra; Labyrinthulomycetes; Thraustochytrida; Thraustochytriaceae; Hondaea
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 11849 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
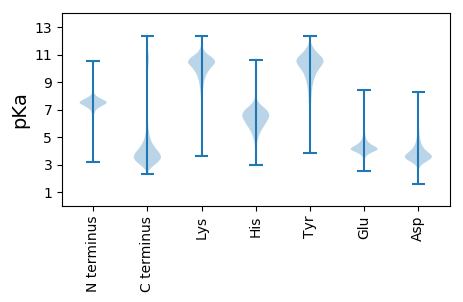
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A2R5G8Q5|A0A2R5G8Q5_9STRA Uncharacterized protein OS=Hondaea fermentalgiana OX=2315210 GN=FCC1311_002622 PE=4 SV=1
MM1 pKa = 6.84FRR3 pKa = 11.84KK4 pKa = 9.32VALVAVLAAACTSAQAQDD22 pKa = 3.4NSDD25 pKa = 3.49WANDD29 pKa = 3.79TMWWDD34 pKa = 3.81DD35 pKa = 4.19DD36 pKa = 4.0EE37 pKa = 5.09PNPVGMLQDD46 pKa = 3.48GDD48 pKa = 4.46LEE50 pKa = 4.45VMYY53 pKa = 10.32EE54 pKa = 3.85ILDD57 pKa = 3.61NDD59 pKa = 4.3MVRR62 pKa = 11.84MTLTYY67 pKa = 7.28PTPNGWIGVGLSDD80 pKa = 4.85DD81 pKa = 3.71GGMVGSHH88 pKa = 5.61AVIGGTGVTGLPAPVGEE105 pKa = 4.62YY106 pKa = 9.86PLEE109 pKa = 4.69SKK111 pKa = 10.92SEE113 pKa = 4.16TPTLYY118 pKa = 10.75SGTNGIYY125 pKa = 8.3NTSVDD130 pKa = 3.74VQDD133 pKa = 4.57SVMTVQFTVEE143 pKa = 4.37SIAGRR148 pKa = 11.84SLDD151 pKa = 4.03LEE153 pKa = 4.64GNGDD157 pKa = 3.7QIIYY161 pKa = 10.29AVYY164 pKa = 10.24RR165 pKa = 11.84GSTWGGAHH173 pKa = 5.74QEE175 pKa = 4.24FGSAMVDD182 pKa = 2.54WSMATPAPTDD192 pKa = 3.59MPSTASRR199 pKa = 11.84AATLSLAAAGMLIGALYY216 pKa = 10.66
MM1 pKa = 6.84FRR3 pKa = 11.84KK4 pKa = 9.32VALVAVLAAACTSAQAQDD22 pKa = 3.4NSDD25 pKa = 3.49WANDD29 pKa = 3.79TMWWDD34 pKa = 3.81DD35 pKa = 4.19DD36 pKa = 4.0EE37 pKa = 5.09PNPVGMLQDD46 pKa = 3.48GDD48 pKa = 4.46LEE50 pKa = 4.45VMYY53 pKa = 10.32EE54 pKa = 3.85ILDD57 pKa = 3.61NDD59 pKa = 4.3MVRR62 pKa = 11.84MTLTYY67 pKa = 7.28PTPNGWIGVGLSDD80 pKa = 4.85DD81 pKa = 3.71GGMVGSHH88 pKa = 5.61AVIGGTGVTGLPAPVGEE105 pKa = 4.62YY106 pKa = 9.86PLEE109 pKa = 4.69SKK111 pKa = 10.92SEE113 pKa = 4.16TPTLYY118 pKa = 10.75SGTNGIYY125 pKa = 8.3NTSVDD130 pKa = 3.74VQDD133 pKa = 4.57SVMTVQFTVEE143 pKa = 4.37SIAGRR148 pKa = 11.84SLDD151 pKa = 4.03LEE153 pKa = 4.64GNGDD157 pKa = 3.7QIIYY161 pKa = 10.29AVYY164 pKa = 10.24RR165 pKa = 11.84GSTWGGAHH173 pKa = 5.74QEE175 pKa = 4.24FGSAMVDD182 pKa = 2.54WSMATPAPTDD192 pKa = 3.59MPSTASRR199 pKa = 11.84AATLSLAAAGMLIGALYY216 pKa = 10.66
Molecular weight: 22.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R5FYP0|A0A2R5FYP0_9STRA CaiB/baiF CoA-transferase family protein DDB_G0269880 OS=Hondaea fermentalgiana OX=2315210 GN=FCC1311_010401 PE=3 SV=1
EEE2 pKa = 4.37RR3 pKa = 11.84AQRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84GLPPAALGRR18 pKa = 11.84CGRR21 pKa = 11.84APRR24 pKa = 11.84AGRR27 pKa = 11.84LRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84AGQRR35 pKa = 11.84HHH37 pKa = 6.18GQALPRR43 pKa = 11.84ALPRR47 pKa = 11.84AARR50 pKa = 11.84DDD52 pKa = 3.17AGAHHH57 pKa = 5.92PRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84AHHH64 pKa = 6.72GAEEE68 pKa = 3.53ALRR71 pKa = 11.84RR72 pKa = 11.84GARR75 pKa = 11.84LLGRR79 pKa = 11.84PRR81 pKa = 11.84GGRR84 pKa = 11.84RR85 pKa = 11.84LRR87 pKa = 11.84ARR89 pKa = 11.84APRR92 pKa = 11.84PAAPAARR99 pKa = 11.84GLRR102 pKa = 11.84ARR104 pKa = 11.84GHHH107 pKa = 6.8GLGLPPRR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84CARR119 pKa = 11.84ARR121 pKa = 11.84GRR123 pKa = 11.84GSGVLV
EEE2 pKa = 4.37RR3 pKa = 11.84AQRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84GLPPAALGRR18 pKa = 11.84CGRR21 pKa = 11.84APRR24 pKa = 11.84AGRR27 pKa = 11.84LRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84AGQRR35 pKa = 11.84HHH37 pKa = 6.18GQALPRR43 pKa = 11.84ALPRR47 pKa = 11.84AARR50 pKa = 11.84DDD52 pKa = 3.17AGAHHH57 pKa = 5.92PRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84AHHH64 pKa = 6.72GAEEE68 pKa = 3.53ALRR71 pKa = 11.84RR72 pKa = 11.84GARR75 pKa = 11.84LLGRR79 pKa = 11.84PRR81 pKa = 11.84GGRR84 pKa = 11.84RR85 pKa = 11.84LRR87 pKa = 11.84ARR89 pKa = 11.84APRR92 pKa = 11.84PAAPAARR99 pKa = 11.84GLRR102 pKa = 11.84ARR104 pKa = 11.84GHHH107 pKa = 6.8GLGLPPRR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84CARR119 pKa = 11.84ARR121 pKa = 11.84GRR123 pKa = 11.84GSGVLV
Molecular weight: 13.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7193445 |
22 |
16137 |
607.1 |
66.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.64 ± 0.025 | 1.481 ± 0.011 |
6.396 ± 0.019 | 7.16 ± 0.024 |
3.562 ± 0.014 | 6.776 ± 0.031 |
2.318 ± 0.01 | 3.865 ± 0.013 |
4.528 ± 0.025 | 9.347 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.223 ± 0.009 | 3.252 ± 0.011 |
4.51 ± 0.017 | 4.049 ± 0.017 |
6.782 ± 0.025 | 7.923 ± 0.03 |
5.216 ± 0.022 | 6.721 ± 0.019 |
1.133 ± 0.007 | 2.12 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
