
Agrobacterium rosae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales;
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
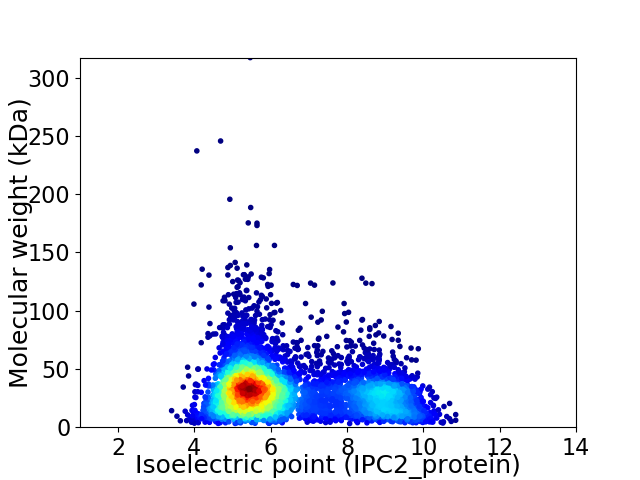
Virtual 2D-PAGE plot for 5403 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
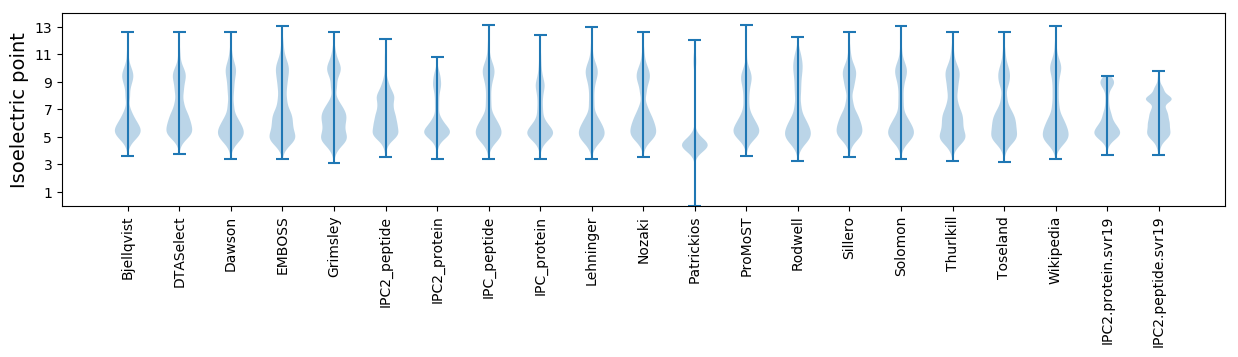
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1R3TD11|A0A1R3TD11_9RHIZ Putative aliphatic sulfonates-binding protein OS=Agrobacterium rosae OX=1972867 GN=ssuA_3 PE=3 SV=1
MM1 pKa = 7.22AQSIMNALGKK11 pKa = 9.22PLYY14 pKa = 10.7YY15 pKa = 10.32SDD17 pKa = 3.55TSTGFFSATGSGPDD31 pKa = 4.28LYY33 pKa = 10.81GTTKK37 pKa = 10.43NDD39 pKa = 3.85SMWGDD44 pKa = 3.3SGVNVTMHH52 pKa = 6.9GGTGDD57 pKa = 4.24DD58 pKa = 3.2IYY60 pKa = 11.69YY61 pKa = 10.45LYY63 pKa = 10.1STINRR68 pKa = 11.84ATEE71 pKa = 3.89AADD74 pKa = 3.79QGTDD78 pKa = 3.98TINTWMSYY86 pKa = 7.36TLPDD90 pKa = 4.13NIEE93 pKa = 3.98NLIVTGDD100 pKa = 3.34GRR102 pKa = 11.84NAFGNALDD110 pKa = 4.69NIISGGSAKK119 pKa = 8.45QTIDD123 pKa = 3.04GGAGNDD129 pKa = 3.45VLTGGGGADD138 pKa = 2.86TFVFAKK144 pKa = 10.91GNGTDD149 pKa = 4.75LITDD153 pKa = 4.62FGSDD157 pKa = 3.59DD158 pKa = 4.75FIRR161 pKa = 11.84LDD163 pKa = 3.88GYY165 pKa = 11.29GVTSFDD171 pKa = 3.95HH172 pKa = 7.08LISTSTQKK180 pKa = 11.11GADD183 pKa = 2.98LWLNFDD189 pKa = 3.62NGEE192 pKa = 4.38SVVLANTTADD202 pKa = 4.54SLDD205 pKa = 3.46ADD207 pKa = 3.98QFRR210 pKa = 11.84LTLDD214 pKa = 3.27RR215 pKa = 11.84SKK217 pKa = 10.57LTSTFADD224 pKa = 3.65EE225 pKa = 4.86FNSLSLSDD233 pKa = 3.82GQSGVWEE240 pKa = 4.37PKK242 pKa = 9.24FWWAGEE248 pKa = 4.19EE249 pKa = 4.31GGSLHH254 pKa = 6.51TNSEE258 pKa = 4.3KK259 pKa = 10.16QWYY262 pKa = 8.9INPAYY267 pKa = 10.42QPTSSVNPFSVQNGVLTITAAATPDD292 pKa = 4.29AISDD296 pKa = 3.67AVNGYY301 pKa = 10.71DD302 pKa = 3.63YY303 pKa = 10.99TSGMLTTHH311 pKa = 6.61SSFSQTYY318 pKa = 9.36GYY320 pKa = 11.18FEE322 pKa = 4.56MRR324 pKa = 11.84ADD326 pKa = 3.65MPSDD330 pKa = 3.24QGAWPAFWLLPEE342 pKa = 4.83DD343 pKa = 4.56GKK345 pKa = 10.23WPPEE349 pKa = 3.66LDD351 pKa = 3.69VIEE354 pKa = 4.62MRR356 pKa = 11.84GQNPNTLILSAHH368 pKa = 5.72SNEE371 pKa = 4.51TGQQTSTINNVNVTSTEE388 pKa = 3.97GFHH391 pKa = 6.86TYY393 pKa = 10.63GLLWDD398 pKa = 4.42EE399 pKa = 5.7DD400 pKa = 4.33HH401 pKa = 6.44ITWYY405 pKa = 10.65FDD407 pKa = 3.53DD408 pKa = 4.08VAVAQIDD415 pKa = 4.38TPTDD419 pKa = 3.25MHH421 pKa = 7.9DD422 pKa = 3.4PMYY425 pKa = 10.14MIVNLAVGGIAGTPTNGLPNGSEE448 pKa = 3.74MKK450 pKa = 9.66IDD452 pKa = 4.06YY453 pKa = 10.26IHH455 pKa = 7.39AYY457 pKa = 9.71SINDD461 pKa = 3.88PAVSAMSQTTSSDD474 pKa = 3.23NLLVV478 pKa = 3.58
MM1 pKa = 7.22AQSIMNALGKK11 pKa = 9.22PLYY14 pKa = 10.7YY15 pKa = 10.32SDD17 pKa = 3.55TSTGFFSATGSGPDD31 pKa = 4.28LYY33 pKa = 10.81GTTKK37 pKa = 10.43NDD39 pKa = 3.85SMWGDD44 pKa = 3.3SGVNVTMHH52 pKa = 6.9GGTGDD57 pKa = 4.24DD58 pKa = 3.2IYY60 pKa = 11.69YY61 pKa = 10.45LYY63 pKa = 10.1STINRR68 pKa = 11.84ATEE71 pKa = 3.89AADD74 pKa = 3.79QGTDD78 pKa = 3.98TINTWMSYY86 pKa = 7.36TLPDD90 pKa = 4.13NIEE93 pKa = 3.98NLIVTGDD100 pKa = 3.34GRR102 pKa = 11.84NAFGNALDD110 pKa = 4.69NIISGGSAKK119 pKa = 8.45QTIDD123 pKa = 3.04GGAGNDD129 pKa = 3.45VLTGGGGADD138 pKa = 2.86TFVFAKK144 pKa = 10.91GNGTDD149 pKa = 4.75LITDD153 pKa = 4.62FGSDD157 pKa = 3.59DD158 pKa = 4.75FIRR161 pKa = 11.84LDD163 pKa = 3.88GYY165 pKa = 11.29GVTSFDD171 pKa = 3.95HH172 pKa = 7.08LISTSTQKK180 pKa = 11.11GADD183 pKa = 2.98LWLNFDD189 pKa = 3.62NGEE192 pKa = 4.38SVVLANTTADD202 pKa = 4.54SLDD205 pKa = 3.46ADD207 pKa = 3.98QFRR210 pKa = 11.84LTLDD214 pKa = 3.27RR215 pKa = 11.84SKK217 pKa = 10.57LTSTFADD224 pKa = 3.65EE225 pKa = 4.86FNSLSLSDD233 pKa = 3.82GQSGVWEE240 pKa = 4.37PKK242 pKa = 9.24FWWAGEE248 pKa = 4.19EE249 pKa = 4.31GGSLHH254 pKa = 6.51TNSEE258 pKa = 4.3KK259 pKa = 10.16QWYY262 pKa = 8.9INPAYY267 pKa = 10.42QPTSSVNPFSVQNGVLTITAAATPDD292 pKa = 4.29AISDD296 pKa = 3.67AVNGYY301 pKa = 10.71DD302 pKa = 3.63YY303 pKa = 10.99TSGMLTTHH311 pKa = 6.61SSFSQTYY318 pKa = 9.36GYY320 pKa = 11.18FEE322 pKa = 4.56MRR324 pKa = 11.84ADD326 pKa = 3.65MPSDD330 pKa = 3.24QGAWPAFWLLPEE342 pKa = 4.83DD343 pKa = 4.56GKK345 pKa = 10.23WPPEE349 pKa = 3.66LDD351 pKa = 3.69VIEE354 pKa = 4.62MRR356 pKa = 11.84GQNPNTLILSAHH368 pKa = 5.72SNEE371 pKa = 4.51TGQQTSTINNVNVTSTEE388 pKa = 3.97GFHH391 pKa = 6.86TYY393 pKa = 10.63GLLWDD398 pKa = 4.42EE399 pKa = 5.7DD400 pKa = 4.33HH401 pKa = 6.44ITWYY405 pKa = 10.65FDD407 pKa = 3.53DD408 pKa = 4.08VAVAQIDD415 pKa = 4.38TPTDD419 pKa = 3.25MHH421 pKa = 7.9DD422 pKa = 3.4PMYY425 pKa = 10.14MIVNLAVGGIAGTPTNGLPNGSEE448 pKa = 3.74MKK450 pKa = 9.66IDD452 pKa = 4.06YY453 pKa = 10.26IHH455 pKa = 7.39AYY457 pKa = 9.71SINDD461 pKa = 3.88PAVSAMSQTTSSDD474 pKa = 3.23NLLVV478 pKa = 3.58
Molecular weight: 51.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1R3U2I2|A0A1R3U2I2_9RHIZ Putative D D-dipeptide transport system permease protein DdpC OS=Agrobacterium rosae OX=1972867 GN=ddpC_8 PE=3 SV=1
MM1 pKa = 7.45LGVHH5 pKa = 6.71LGVFFNKK12 pKa = 9.25VFSAHH17 pKa = 4.98VALAARR23 pKa = 11.84RR24 pKa = 11.84ARR26 pKa = 11.84AGSVGLNSFGRR37 pKa = 11.84FAKK40 pKa = 9.93WISASIMPPPSLVSARR56 pKa = 3.78
MM1 pKa = 7.45LGVHH5 pKa = 6.71LGVFFNKK12 pKa = 9.25VFSAHH17 pKa = 4.98VALAARR23 pKa = 11.84RR24 pKa = 11.84ARR26 pKa = 11.84AGSVGLNSFGRR37 pKa = 11.84FAKK40 pKa = 9.93WISASIMPPPSLVSARR56 pKa = 3.78
Molecular weight: 5.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1650202 |
29 |
2830 |
305.4 |
33.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.176 ± 0.041 | 0.768 ± 0.009 |
5.777 ± 0.025 | 5.713 ± 0.031 |
3.971 ± 0.022 | 7.986 ± 0.032 |
2.03 ± 0.016 | 5.929 ± 0.026 |
4.104 ± 0.029 | 9.816 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.623 ± 0.015 | 3.1 ± 0.018 |
4.687 ± 0.023 | 3.313 ± 0.02 |
6.283 ± 0.03 | 6.199 ± 0.026 |
5.614 ± 0.022 | 7.314 ± 0.026 |
1.257 ± 0.014 | 2.341 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
