
Herbiconiux ginsengi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia;
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
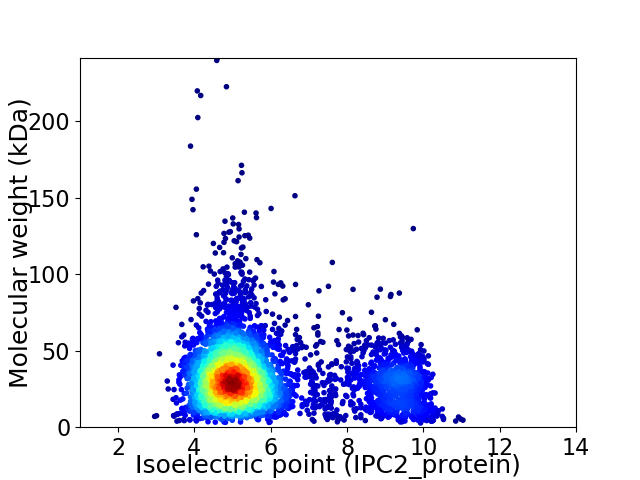
Virtual 2D-PAGE plot for 4507 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3TJS6|A0A1H3TJS6_9MICO ATP-binding cassette subfamily B OS=Herbiconiux ginsengi OX=381665 GN=SAMN05216554_4275 PE=4 SV=1
MM1 pKa = 7.62LANKK5 pKa = 10.0SFTLIGSGVSVMLIAALTGCSASGGVPGASKK36 pKa = 10.77GGEE39 pKa = 3.9LRR41 pKa = 11.84LALLGDD47 pKa = 4.14ISTPDD52 pKa = 3.95PDD54 pKa = 3.53TAYY57 pKa = 10.64DD58 pKa = 3.79GSEE61 pKa = 3.96LNIVNSAYY69 pKa = 10.05EE70 pKa = 3.98GLLGYY75 pKa = 10.45VPGKK79 pKa = 9.5QEE81 pKa = 3.93PEE83 pKa = 4.06LTGVLATEE91 pKa = 4.85WTASANNTVFTFRR104 pKa = 11.84LRR106 pKa = 11.84EE107 pKa = 3.85GVTFHH112 pKa = 7.72DD113 pKa = 4.45GTPFTADD120 pKa = 4.25AIQSSFDD127 pKa = 3.13RR128 pKa = 11.84RR129 pKa = 11.84NAIGEE134 pKa = 4.41GPSYY138 pKa = 8.43MTAGVASVVAADD150 pKa = 4.66DD151 pKa = 4.13FNVTITLRR159 pKa = 11.84QPNSSFLDD167 pKa = 3.65LLASPFGPKK176 pKa = 9.23MISPTALKK184 pKa = 9.39EE185 pKa = 4.07HH186 pKa = 6.6PVVNGSEE193 pKa = 3.96DD194 pKa = 3.21WFDD197 pKa = 4.53SNDD200 pKa = 3.68AGTGPYY206 pKa = 9.57TYY208 pKa = 10.94GAFAPGTSYY217 pKa = 11.2QLNAYY222 pKa = 8.74GGYY225 pKa = 9.23WGDD228 pKa = 3.43APGYY232 pKa = 9.65DD233 pKa = 2.96AVSFDD238 pKa = 3.83VFSNSATIQLEE249 pKa = 4.22LQDD252 pKa = 4.43GGVDD256 pKa = 3.77GLIGYY261 pKa = 10.14ADD263 pKa = 4.19ASTFDD268 pKa = 3.14QLKK271 pKa = 10.51RR272 pKa = 11.84SDD274 pKa = 3.96RR275 pKa = 11.84LSAYY279 pKa = 9.32AFPSMEE285 pKa = 4.26TPTMFVNPQSPDD297 pKa = 3.2LADD300 pKa = 4.26PDD302 pKa = 3.7VRR304 pKa = 11.84RR305 pKa = 11.84SFISSIDD312 pKa = 3.62FNALADD318 pKa = 3.67TALADD323 pKa = 3.81TATPTDD329 pKa = 3.64GVFPTTLLPANLNTQAITYY348 pKa = 8.88NPNALAQLAKK358 pKa = 10.36GQLAGSTITIAYY370 pKa = 8.0AQAGPAAQALSDD382 pKa = 3.79NLAAVLNGAGIAAEE396 pKa = 4.16SVGYY400 pKa = 8.33DD401 pKa = 3.01TGSYY405 pKa = 9.62YY406 pKa = 11.06AALTEE411 pKa = 4.46GAAAPDD417 pKa = 3.2ITFYY421 pKa = 11.31SGFPDD426 pKa = 3.73TASPEE431 pKa = 3.25AWAFVFYY438 pKa = 10.37TPAGGLDD445 pKa = 3.73LFGADD450 pKa = 3.53VPGVADD456 pKa = 6.23LIDD459 pKa = 3.47QATVTGDD466 pKa = 3.22DD467 pKa = 3.56ALYY470 pKa = 10.88GDD472 pKa = 3.85VAQRR476 pKa = 11.84VSEE479 pKa = 4.13SGYY482 pKa = 7.69WRR484 pKa = 11.84SVATSLGLAVFQKK497 pKa = 10.83GISGVDD503 pKa = 2.94RR504 pKa = 11.84AYY506 pKa = 11.24SPVITGILDD515 pKa = 3.23IALIEE520 pKa = 4.34PPAA523 pKa = 5.46
MM1 pKa = 7.62LANKK5 pKa = 10.0SFTLIGSGVSVMLIAALTGCSASGGVPGASKK36 pKa = 10.77GGEE39 pKa = 3.9LRR41 pKa = 11.84LALLGDD47 pKa = 4.14ISTPDD52 pKa = 3.95PDD54 pKa = 3.53TAYY57 pKa = 10.64DD58 pKa = 3.79GSEE61 pKa = 3.96LNIVNSAYY69 pKa = 10.05EE70 pKa = 3.98GLLGYY75 pKa = 10.45VPGKK79 pKa = 9.5QEE81 pKa = 3.93PEE83 pKa = 4.06LTGVLATEE91 pKa = 4.85WTASANNTVFTFRR104 pKa = 11.84LRR106 pKa = 11.84EE107 pKa = 3.85GVTFHH112 pKa = 7.72DD113 pKa = 4.45GTPFTADD120 pKa = 4.25AIQSSFDD127 pKa = 3.13RR128 pKa = 11.84RR129 pKa = 11.84NAIGEE134 pKa = 4.41GPSYY138 pKa = 8.43MTAGVASVVAADD150 pKa = 4.66DD151 pKa = 4.13FNVTITLRR159 pKa = 11.84QPNSSFLDD167 pKa = 3.65LLASPFGPKK176 pKa = 9.23MISPTALKK184 pKa = 9.39EE185 pKa = 4.07HH186 pKa = 6.6PVVNGSEE193 pKa = 3.96DD194 pKa = 3.21WFDD197 pKa = 4.53SNDD200 pKa = 3.68AGTGPYY206 pKa = 9.57TYY208 pKa = 10.94GAFAPGTSYY217 pKa = 11.2QLNAYY222 pKa = 8.74GGYY225 pKa = 9.23WGDD228 pKa = 3.43APGYY232 pKa = 9.65DD233 pKa = 2.96AVSFDD238 pKa = 3.83VFSNSATIQLEE249 pKa = 4.22LQDD252 pKa = 4.43GGVDD256 pKa = 3.77GLIGYY261 pKa = 10.14ADD263 pKa = 4.19ASTFDD268 pKa = 3.14QLKK271 pKa = 10.51RR272 pKa = 11.84SDD274 pKa = 3.96RR275 pKa = 11.84LSAYY279 pKa = 9.32AFPSMEE285 pKa = 4.26TPTMFVNPQSPDD297 pKa = 3.2LADD300 pKa = 4.26PDD302 pKa = 3.7VRR304 pKa = 11.84RR305 pKa = 11.84SFISSIDD312 pKa = 3.62FNALADD318 pKa = 3.67TALADD323 pKa = 3.81TATPTDD329 pKa = 3.64GVFPTTLLPANLNTQAITYY348 pKa = 8.88NPNALAQLAKK358 pKa = 10.36GQLAGSTITIAYY370 pKa = 8.0AQAGPAAQALSDD382 pKa = 3.79NLAAVLNGAGIAAEE396 pKa = 4.16SVGYY400 pKa = 8.33DD401 pKa = 3.01TGSYY405 pKa = 9.62YY406 pKa = 11.06AALTEE411 pKa = 4.46GAAAPDD417 pKa = 3.2ITFYY421 pKa = 11.31SGFPDD426 pKa = 3.73TASPEE431 pKa = 3.25AWAFVFYY438 pKa = 10.37TPAGGLDD445 pKa = 3.73LFGADD450 pKa = 3.53VPGVADD456 pKa = 6.23LIDD459 pKa = 3.47QATVTGDD466 pKa = 3.22DD467 pKa = 3.56ALYY470 pKa = 10.88GDD472 pKa = 3.85VAQRR476 pKa = 11.84VSEE479 pKa = 4.13SGYY482 pKa = 7.69WRR484 pKa = 11.84SVATSLGLAVFQKK497 pKa = 10.83GISGVDD503 pKa = 2.94RR504 pKa = 11.84AYY506 pKa = 11.24SPVITGILDD515 pKa = 3.23IALIEE520 pKa = 4.34PPAA523 pKa = 5.46
Molecular weight: 54.22 kDa
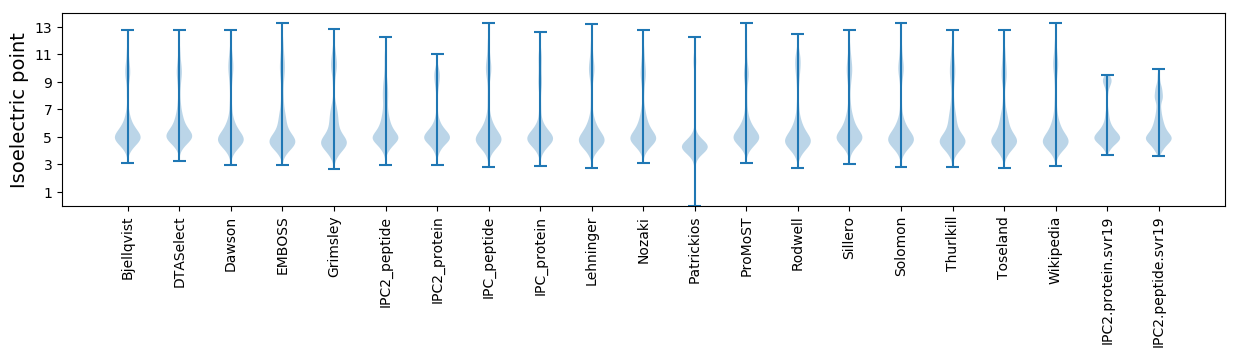
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H3TLW1|A0A1H3TLW1_9MICO AraC-type DNA-binding protein OS=Herbiconiux ginsengi OX=381665 GN=SAMN05216554_4304 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 10.23VRR4 pKa = 11.84NSLGSMKK11 pKa = 10.21KK12 pKa = 7.81MQGSQVVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84TFVINHH30 pKa = 6.31ANPRR34 pKa = 11.84FKK36 pKa = 10.81ARR38 pKa = 11.84QGG40 pKa = 3.28
MM1 pKa = 7.63KK2 pKa = 10.23VRR4 pKa = 11.84NSLGSMKK11 pKa = 10.21KK12 pKa = 7.81MQGSQVVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84TFVINHH30 pKa = 6.31ANPRR34 pKa = 11.84FKK36 pKa = 10.81ARR38 pKa = 11.84QGG40 pKa = 3.28
Molecular weight: 4.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1471252 |
25 |
2301 |
326.4 |
34.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.319 ± 0.05 | 0.479 ± 0.009 |
6.157 ± 0.029 | 5.451 ± 0.037 |
3.248 ± 0.022 | 9.043 ± 0.038 |
1.967 ± 0.02 | 4.625 ± 0.028 |
1.925 ± 0.023 | 9.999 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.657 ± 0.013 | 2.138 ± 0.022 |
5.531 ± 0.028 | 2.721 ± 0.021 |
6.872 ± 0.044 | 6.181 ± 0.026 |
6.275 ± 0.04 | 8.906 ± 0.038 |
1.46 ± 0.016 | 2.046 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
