
Changjiang tombus-like virus 14
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.41
Get precalculated fractions of proteins
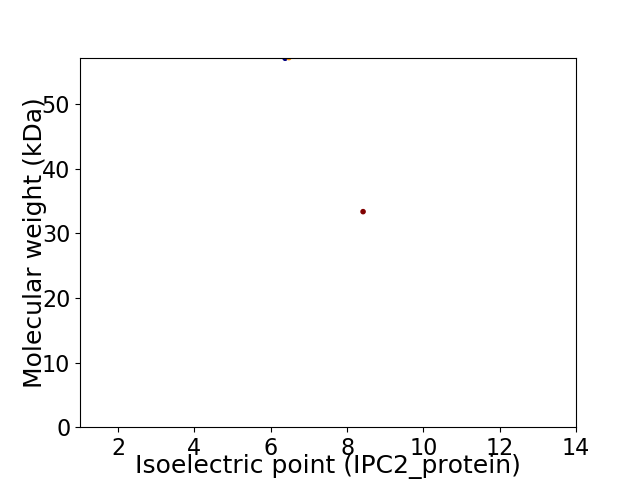
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG69|A0A1L3KG69_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 14 OX=1922807 PE=4 SV=1
MM1 pKa = 7.93DD2 pKa = 4.68RR3 pKa = 11.84RR4 pKa = 11.84MPTPGIVYY12 pKa = 7.51THH14 pKa = 6.54NNSLDD19 pKa = 3.41NVLRR23 pKa = 11.84GIGEE27 pKa = 4.21RR28 pKa = 11.84LKK30 pKa = 10.37MVSDD34 pKa = 3.85GRR36 pKa = 11.84GGFCFPPRR44 pKa = 11.84PKK46 pKa = 10.65VFDD49 pKa = 3.78LEE51 pKa = 4.56EE52 pKa = 3.96YY53 pKa = 10.37RR54 pKa = 11.84SRR56 pKa = 11.84LLRR59 pKa = 11.84KK60 pKa = 8.68MPKK63 pKa = 9.29FDD65 pKa = 4.72APITLDD71 pKa = 3.87EE72 pKa = 4.58FVQLYY77 pKa = 10.1DD78 pKa = 3.32GPKK81 pKa = 8.26RR82 pKa = 11.84KK83 pKa = 9.53RR84 pKa = 11.84YY85 pKa = 9.05EE86 pKa = 4.0SAVEE90 pKa = 3.66QLVRR94 pKa = 11.84DD95 pKa = 4.2GLQPIDD101 pKa = 3.81GDD103 pKa = 3.26IALFIKK109 pKa = 10.34DD110 pKa = 4.15EE111 pKa = 5.04KK112 pKa = 9.69ICSWSKK118 pKa = 10.5VDD120 pKa = 3.71PAPRR124 pKa = 11.84LISPRR129 pKa = 11.84SPKK132 pKa = 10.47YY133 pKa = 9.55CVQLGRR139 pKa = 11.84YY140 pKa = 7.8IKK142 pKa = 10.23PIEE145 pKa = 4.26HH146 pKa = 7.24LLYY149 pKa = 10.69KK150 pKa = 10.52AVARR154 pKa = 11.84VWGEE158 pKa = 3.6TTIAKK163 pKa = 9.78GLNFNEE169 pKa = 4.14RR170 pKa = 11.84GVLIQQKK177 pKa = 8.65WEE179 pKa = 4.15SFNDD183 pKa = 3.85PVAVGLDD190 pKa = 3.17ASRR193 pKa = 11.84FDD195 pKa = 3.44QHH197 pKa = 7.91VSEE200 pKa = 4.87LALMWEE206 pKa = 4.13HH207 pKa = 5.91SVYY210 pKa = 9.29MRR212 pKa = 11.84CYY214 pKa = 9.85PRR216 pKa = 11.84EE217 pKa = 3.94EE218 pKa = 4.44CDD220 pKa = 3.23GKK222 pKa = 10.57LARR225 pKa = 11.84LLEE228 pKa = 4.35RR229 pKa = 11.84QLVNRR234 pKa = 11.84GRR236 pKa = 11.84CYY238 pKa = 10.77VEE240 pKa = 3.6NHH242 pKa = 5.06LVEE245 pKa = 4.34YY246 pKa = 7.64EE247 pKa = 4.05HH248 pKa = 7.23RR249 pKa = 11.84GGRR252 pKa = 11.84MSGDD256 pKa = 3.3MNTALGNCLIMTGLVWEE273 pKa = 5.01HH274 pKa = 5.81ARR276 pKa = 11.84QLGVTVKK283 pKa = 10.74LINDD287 pKa = 3.59GDD289 pKa = 4.07DD290 pKa = 3.08CVVFMEE296 pKa = 4.4RR297 pKa = 11.84ADD299 pKa = 3.65LARR302 pKa = 11.84YY303 pKa = 9.74LDD305 pKa = 4.45GLEE308 pKa = 3.53EE309 pKa = 4.07WFRR312 pKa = 11.84ARR314 pKa = 11.84GFTMKK319 pKa = 10.2VEE321 pKa = 4.28KK322 pKa = 9.89PAFEE326 pKa = 4.46LEE328 pKa = 4.12QIEE331 pKa = 4.71FCQCRR336 pKa = 11.84PVWNGEE342 pKa = 3.9QYY344 pKa = 10.35TMCRR348 pKa = 11.84NVFKK352 pKa = 10.97ALFTDD357 pKa = 5.38GVHH360 pKa = 6.26VGRR363 pKa = 11.84TLSEE367 pKa = 3.79IQHH370 pKa = 5.87IRR372 pKa = 11.84SATSKK377 pKa = 10.58CGKK380 pKa = 9.1VWSKK384 pKa = 10.76GLPIFGEE391 pKa = 4.6FYY393 pKa = 11.05EE394 pKa = 4.59FLSCEE399 pKa = 4.02APRR402 pKa = 11.84GKK404 pKa = 10.9SNFYY408 pKa = 10.22GDD410 pKa = 3.92YY411 pKa = 9.75RR412 pKa = 11.84HH413 pKa = 7.03SGTVWQAKK421 pKa = 8.83GCVSGTTHH429 pKa = 5.91ITDD432 pKa = 3.25EE433 pKa = 4.34ARR435 pKa = 11.84ASFHH439 pKa = 6.4RR440 pKa = 11.84AFGITGSEE448 pKa = 3.93QVLVEE453 pKa = 5.0DD454 pKa = 5.81FYY456 pKa = 12.1KK457 pKa = 10.79DD458 pKa = 3.86LPKK461 pKa = 8.72TTYY464 pKa = 7.68DD465 pKa = 3.62TPQDD469 pKa = 3.39ALVYY473 pKa = 9.98NPQISPDD480 pKa = 3.74SYY482 pKa = 10.59PLFVSEE488 pKa = 4.64SLCEE492 pKa = 3.83IVFNKK497 pKa = 10.3
MM1 pKa = 7.93DD2 pKa = 4.68RR3 pKa = 11.84RR4 pKa = 11.84MPTPGIVYY12 pKa = 7.51THH14 pKa = 6.54NNSLDD19 pKa = 3.41NVLRR23 pKa = 11.84GIGEE27 pKa = 4.21RR28 pKa = 11.84LKK30 pKa = 10.37MVSDD34 pKa = 3.85GRR36 pKa = 11.84GGFCFPPRR44 pKa = 11.84PKK46 pKa = 10.65VFDD49 pKa = 3.78LEE51 pKa = 4.56EE52 pKa = 3.96YY53 pKa = 10.37RR54 pKa = 11.84SRR56 pKa = 11.84LLRR59 pKa = 11.84KK60 pKa = 8.68MPKK63 pKa = 9.29FDD65 pKa = 4.72APITLDD71 pKa = 3.87EE72 pKa = 4.58FVQLYY77 pKa = 10.1DD78 pKa = 3.32GPKK81 pKa = 8.26RR82 pKa = 11.84KK83 pKa = 9.53RR84 pKa = 11.84YY85 pKa = 9.05EE86 pKa = 4.0SAVEE90 pKa = 3.66QLVRR94 pKa = 11.84DD95 pKa = 4.2GLQPIDD101 pKa = 3.81GDD103 pKa = 3.26IALFIKK109 pKa = 10.34DD110 pKa = 4.15EE111 pKa = 5.04KK112 pKa = 9.69ICSWSKK118 pKa = 10.5VDD120 pKa = 3.71PAPRR124 pKa = 11.84LISPRR129 pKa = 11.84SPKK132 pKa = 10.47YY133 pKa = 9.55CVQLGRR139 pKa = 11.84YY140 pKa = 7.8IKK142 pKa = 10.23PIEE145 pKa = 4.26HH146 pKa = 7.24LLYY149 pKa = 10.69KK150 pKa = 10.52AVARR154 pKa = 11.84VWGEE158 pKa = 3.6TTIAKK163 pKa = 9.78GLNFNEE169 pKa = 4.14RR170 pKa = 11.84GVLIQQKK177 pKa = 8.65WEE179 pKa = 4.15SFNDD183 pKa = 3.85PVAVGLDD190 pKa = 3.17ASRR193 pKa = 11.84FDD195 pKa = 3.44QHH197 pKa = 7.91VSEE200 pKa = 4.87LALMWEE206 pKa = 4.13HH207 pKa = 5.91SVYY210 pKa = 9.29MRR212 pKa = 11.84CYY214 pKa = 9.85PRR216 pKa = 11.84EE217 pKa = 3.94EE218 pKa = 4.44CDD220 pKa = 3.23GKK222 pKa = 10.57LARR225 pKa = 11.84LLEE228 pKa = 4.35RR229 pKa = 11.84QLVNRR234 pKa = 11.84GRR236 pKa = 11.84CYY238 pKa = 10.77VEE240 pKa = 3.6NHH242 pKa = 5.06LVEE245 pKa = 4.34YY246 pKa = 7.64EE247 pKa = 4.05HH248 pKa = 7.23RR249 pKa = 11.84GGRR252 pKa = 11.84MSGDD256 pKa = 3.3MNTALGNCLIMTGLVWEE273 pKa = 5.01HH274 pKa = 5.81ARR276 pKa = 11.84QLGVTVKK283 pKa = 10.74LINDD287 pKa = 3.59GDD289 pKa = 4.07DD290 pKa = 3.08CVVFMEE296 pKa = 4.4RR297 pKa = 11.84ADD299 pKa = 3.65LARR302 pKa = 11.84YY303 pKa = 9.74LDD305 pKa = 4.45GLEE308 pKa = 3.53EE309 pKa = 4.07WFRR312 pKa = 11.84ARR314 pKa = 11.84GFTMKK319 pKa = 10.2VEE321 pKa = 4.28KK322 pKa = 9.89PAFEE326 pKa = 4.46LEE328 pKa = 4.12QIEE331 pKa = 4.71FCQCRR336 pKa = 11.84PVWNGEE342 pKa = 3.9QYY344 pKa = 10.35TMCRR348 pKa = 11.84NVFKK352 pKa = 10.97ALFTDD357 pKa = 5.38GVHH360 pKa = 6.26VGRR363 pKa = 11.84TLSEE367 pKa = 3.79IQHH370 pKa = 5.87IRR372 pKa = 11.84SATSKK377 pKa = 10.58CGKK380 pKa = 9.1VWSKK384 pKa = 10.76GLPIFGEE391 pKa = 4.6FYY393 pKa = 11.05EE394 pKa = 4.59FLSCEE399 pKa = 4.02APRR402 pKa = 11.84GKK404 pKa = 10.9SNFYY408 pKa = 10.22GDD410 pKa = 3.92YY411 pKa = 9.75RR412 pKa = 11.84HH413 pKa = 7.03SGTVWQAKK421 pKa = 8.83GCVSGTTHH429 pKa = 5.91ITDD432 pKa = 3.25EE433 pKa = 4.34ARR435 pKa = 11.84ASFHH439 pKa = 6.4RR440 pKa = 11.84AFGITGSEE448 pKa = 3.93QVLVEE453 pKa = 5.0DD454 pKa = 5.81FYY456 pKa = 12.1KK457 pKa = 10.79DD458 pKa = 3.86LPKK461 pKa = 8.72TTYY464 pKa = 7.68DD465 pKa = 3.62TPQDD469 pKa = 3.39ALVYY473 pKa = 9.98NPQISPDD480 pKa = 3.74SYY482 pKa = 10.59PLFVSEE488 pKa = 4.64SLCEE492 pKa = 3.83IVFNKK497 pKa = 10.3
Molecular weight: 57.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG69|A0A1L3KG69_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 14 OX=1922807 PE=4 SV=1
MM1 pKa = 7.8AKK3 pKa = 9.49TKK5 pKa = 10.21VKK7 pKa = 10.1KK8 pKa = 10.66VKK10 pKa = 10.52ASSQYY15 pKa = 7.57QRR17 pKa = 11.84KK18 pKa = 6.01MTGMYY23 pKa = 9.59KK24 pKa = 10.01QLEE27 pKa = 4.25LGKK30 pKa = 10.01LDD32 pKa = 4.84KK33 pKa = 10.79EE34 pKa = 4.39ALQAAAMYY42 pKa = 10.26ADD44 pKa = 4.08PCGADD49 pKa = 4.07LVPSVYY55 pKa = 10.43PGDD58 pKa = 3.22RR59 pKa = 11.84GFINRR64 pKa = 11.84FTNYY68 pKa = 8.37NTVGNGATEE77 pKa = 4.17TCAIMIWRR85 pKa = 11.84PGTQGLYY92 pKa = 9.49QAGAVTSATTAAVTYY107 pKa = 7.95GTFFPGLPFLNTNASKK123 pKa = 9.41TRR125 pKa = 11.84CAAFCATVRR134 pKa = 11.84PVSAPNTATGTIYY147 pKa = 10.63FGIVNARR154 pKa = 11.84AVRR157 pKa = 11.84SGATLSPNDD166 pKa = 4.09YY167 pKa = 10.6ISLCSEE173 pKa = 4.31SVSCSQALMAPLEE186 pKa = 4.43VRR188 pKa = 11.84WSPGDD193 pKa = 3.19LDD195 pKa = 3.79ARR197 pKa = 11.84YY198 pKa = 10.41SEE200 pKa = 4.45VNNPNVAADD209 pKa = 3.77DD210 pKa = 3.49SDD212 pKa = 4.8YY213 pKa = 11.75NVLIVVAIGIPANTGMQFRR232 pKa = 11.84ATAVMEE238 pKa = 4.25WAPNANIGITNDD250 pKa = 3.14ATSVKK255 pKa = 10.12PSACDD260 pKa = 3.34KK261 pKa = 10.51EE262 pKa = 5.15CVINYY267 pKa = 8.09LKK269 pKa = 10.91KK270 pKa = 10.04KK271 pKa = 10.57DD272 pKa = 4.03RR273 pKa = 11.84DD274 pKa = 3.32WWWNLGKK281 pKa = 8.92KK282 pKa = 7.03TLNVGKK288 pKa = 10.25SVVQGYY294 pKa = 6.8YY295 pKa = 9.47TGGAIGAMGSLLKK308 pKa = 10.33YY309 pKa = 10.74AKK311 pKa = 10.41
MM1 pKa = 7.8AKK3 pKa = 9.49TKK5 pKa = 10.21VKK7 pKa = 10.1KK8 pKa = 10.66VKK10 pKa = 10.52ASSQYY15 pKa = 7.57QRR17 pKa = 11.84KK18 pKa = 6.01MTGMYY23 pKa = 9.59KK24 pKa = 10.01QLEE27 pKa = 4.25LGKK30 pKa = 10.01LDD32 pKa = 4.84KK33 pKa = 10.79EE34 pKa = 4.39ALQAAAMYY42 pKa = 10.26ADD44 pKa = 4.08PCGADD49 pKa = 4.07LVPSVYY55 pKa = 10.43PGDD58 pKa = 3.22RR59 pKa = 11.84GFINRR64 pKa = 11.84FTNYY68 pKa = 8.37NTVGNGATEE77 pKa = 4.17TCAIMIWRR85 pKa = 11.84PGTQGLYY92 pKa = 9.49QAGAVTSATTAAVTYY107 pKa = 7.95GTFFPGLPFLNTNASKK123 pKa = 9.41TRR125 pKa = 11.84CAAFCATVRR134 pKa = 11.84PVSAPNTATGTIYY147 pKa = 10.63FGIVNARR154 pKa = 11.84AVRR157 pKa = 11.84SGATLSPNDD166 pKa = 4.09YY167 pKa = 10.6ISLCSEE173 pKa = 4.31SVSCSQALMAPLEE186 pKa = 4.43VRR188 pKa = 11.84WSPGDD193 pKa = 3.19LDD195 pKa = 3.79ARR197 pKa = 11.84YY198 pKa = 10.41SEE200 pKa = 4.45VNNPNVAADD209 pKa = 3.77DD210 pKa = 3.49SDD212 pKa = 4.8YY213 pKa = 11.75NVLIVVAIGIPANTGMQFRR232 pKa = 11.84ATAVMEE238 pKa = 4.25WAPNANIGITNDD250 pKa = 3.14ATSVKK255 pKa = 10.12PSACDD260 pKa = 3.34KK261 pKa = 10.51EE262 pKa = 5.15CVINYY267 pKa = 8.09LKK269 pKa = 10.91KK270 pKa = 10.04KK271 pKa = 10.57DD272 pKa = 4.03RR273 pKa = 11.84DD274 pKa = 3.32WWWNLGKK281 pKa = 8.92KK282 pKa = 7.03TLNVGKK288 pKa = 10.25SVVQGYY294 pKa = 6.8YY295 pKa = 9.47TGGAIGAMGSLLKK308 pKa = 10.33YY309 pKa = 10.74AKK311 pKa = 10.41
Molecular weight: 33.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
808 |
311 |
497 |
404.0 |
45.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.045 ± 2.748 | 2.847 ± 0.156 |
5.446 ± 0.539 | 5.569 ± 1.71 |
4.084 ± 0.862 | 7.921 ± 0.251 |
1.485 ± 0.847 | 4.332 ± 0.086 |
5.817 ± 0.35 | 7.673 ± 0.892 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.599 ± 0.168 | 4.455 ± 1.127 |
4.95 ± 0.073 | 3.218 ± 0.185 |
6.312 ± 1.4 | 5.817 ± 0.35 |
5.693 ± 1.338 | 7.55 ± 0.096 |
1.856 ± 0.042 | 4.332 ± 0.28 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
