
Candidatus Saccharimonas aalborgensis
Taxonomy: cellular organisms; Bacteria; Bacteria incertae sedis; Bacteria candidate phyla; Candidatus Saccharibacteria; Candidatus Saccharimonas
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
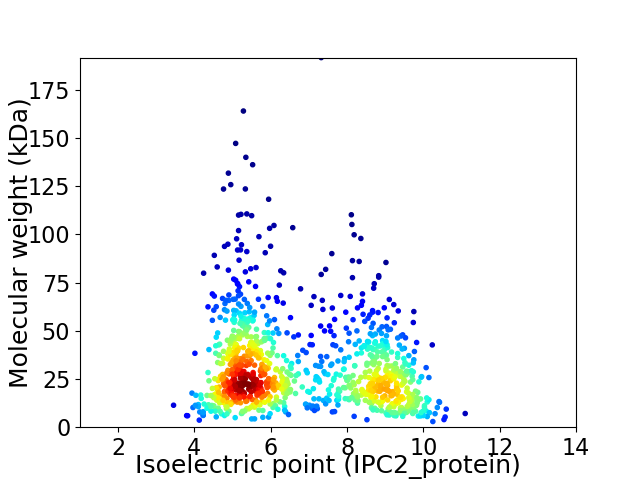
Virtual 2D-PAGE plot for 1033 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R4PW32|R4PW32_9BACT Peptide chain release factor 2 OS=Candidatus Saccharimonas aalborgensis OX=1332188 GN=prfB PE=3 SV=1
MM1 pKa = 7.37TMQTIKK7 pKa = 10.62RR8 pKa = 11.84VVLSLVIAAAPFFVVSPVSAAVDD31 pKa = 3.44TCTWTGAVDD40 pKa = 3.86NTWSNGGNWTGCDD53 pKa = 3.14NAGVPEE59 pKa = 4.59NGDD62 pKa = 3.29ALTFPEE68 pKa = 4.28VANNKK73 pKa = 9.75SMNNDD78 pKa = 3.25LVGLSVTGLSFTGTGYY94 pKa = 8.83TVGGNAFSISTVTALDD110 pKa = 3.25ATEE113 pKa = 4.4SVTINADD120 pKa = 2.77ITYY123 pKa = 8.52TAFNASITPSAGKK136 pKa = 7.82TVTINGVSNFTLGGGEE152 pKa = 4.18VNIGGSGRR160 pKa = 11.84SGTVDD165 pKa = 3.9FYY167 pKa = 12.08GNITGTAAQLVAVSGATAVVRR188 pKa = 11.84GAANTYY194 pKa = 8.19TAATVGAEE202 pKa = 4.22SNAHH206 pKa = 5.24FVCRR210 pKa = 11.84SLTCFGDD217 pKa = 3.56NANDD221 pKa = 3.5IYY223 pKa = 11.42SGGGVVEE230 pKa = 4.23LHH232 pKa = 5.16TTGTYY237 pKa = 10.39VNNIQTSVVTPDD249 pKa = 3.35TSSIWAYY256 pKa = 11.25DD257 pKa = 3.55DD258 pKa = 3.4VTLNGAITVNDD269 pKa = 3.8GLYY272 pKa = 10.47FGQGTNGKK280 pKa = 7.1TLAVDD285 pKa = 3.92GDD287 pKa = 4.11VNLAAEE293 pKa = 4.74DD294 pKa = 3.87LQISGMSRR302 pKa = 11.84AANVVINGVVSGNHH316 pKa = 6.39SIFVYY321 pKa = 10.73SSTLTLAGTNTYY333 pKa = 9.57TGATYY338 pKa = 8.2ATSNNATIEE347 pKa = 4.17VTNQSGLGTSGAGTQIGDD365 pKa = 3.62GDD367 pKa = 3.92SLEE370 pKa = 4.76FNFASAQTVSEE381 pKa = 4.16PLYY384 pKa = 11.22VMGTGVSGTGAVVQTGASTTLSGAIALAGDD414 pKa = 3.58TTFGVDD420 pKa = 3.43SNALFSAFMLSGVISGTGNITVTTSPTTVVANSSIQFTGASPNTYY465 pKa = 8.79TGKK468 pKa = 8.82LTVQGVRR475 pKa = 11.84FYY477 pKa = 10.19PSKK480 pKa = 10.13AASVVAVTGDD490 pKa = 3.38MDD492 pKa = 4.11VLASASKK499 pKa = 10.15PSTVEE504 pKa = 3.72TAFSEE509 pKa = 4.82SIADD513 pKa = 3.63TSHH516 pKa = 4.56VHH518 pKa = 5.85LVKK521 pKa = 10.88DD522 pKa = 4.34GSNKK526 pKa = 9.89ASLSIGSSATEE537 pKa = 3.96TIGYY541 pKa = 6.74VTGDD545 pKa = 3.56GEE547 pKa = 4.88LSVGANGAAVVLTSNADD564 pKa = 3.37YY565 pKa = 9.86TFSGTVSKK573 pKa = 10.5FSNFPAGDD581 pKa = 3.56SFVVKK586 pKa = 10.32KK587 pKa = 9.82GTGTATLTGGLDD599 pKa = 3.18TSYY602 pKa = 10.72IAGVAPVFGAQEE614 pKa = 3.97GSLILNGVFTEE625 pKa = 4.78AGTQVLSGATLKK637 pKa = 10.25GTGTVGAVTVDD648 pKa = 2.85NGGAVSVGNSPGCMTFGSLTLNSGSTYY675 pKa = 7.16TQEE678 pKa = 3.95ISGTTACTQYY688 pKa = 11.4DD689 pKa = 3.67QATVTGAAVLGNATLATTLSTNPADD714 pKa = 3.54GTVFTILTAASVSGTFNGLPDD735 pKa = 4.0GATVNVNGVTLRR747 pKa = 11.84INYY750 pKa = 7.47TATSVTLTKK759 pKa = 10.73VGGNLAPTGSSTAIYY774 pKa = 10.08GLLGLCMVLLSTLALRR790 pKa = 11.84RR791 pKa = 11.84YY792 pKa = 9.74RR793 pKa = 11.84FNNHH797 pKa = 4.62TNN799 pKa = 3.14
MM1 pKa = 7.37TMQTIKK7 pKa = 10.62RR8 pKa = 11.84VVLSLVIAAAPFFVVSPVSAAVDD31 pKa = 3.44TCTWTGAVDD40 pKa = 3.86NTWSNGGNWTGCDD53 pKa = 3.14NAGVPEE59 pKa = 4.59NGDD62 pKa = 3.29ALTFPEE68 pKa = 4.28VANNKK73 pKa = 9.75SMNNDD78 pKa = 3.25LVGLSVTGLSFTGTGYY94 pKa = 8.83TVGGNAFSISTVTALDD110 pKa = 3.25ATEE113 pKa = 4.4SVTINADD120 pKa = 2.77ITYY123 pKa = 8.52TAFNASITPSAGKK136 pKa = 7.82TVTINGVSNFTLGGGEE152 pKa = 4.18VNIGGSGRR160 pKa = 11.84SGTVDD165 pKa = 3.9FYY167 pKa = 12.08GNITGTAAQLVAVSGATAVVRR188 pKa = 11.84GAANTYY194 pKa = 8.19TAATVGAEE202 pKa = 4.22SNAHH206 pKa = 5.24FVCRR210 pKa = 11.84SLTCFGDD217 pKa = 3.56NANDD221 pKa = 3.5IYY223 pKa = 11.42SGGGVVEE230 pKa = 4.23LHH232 pKa = 5.16TTGTYY237 pKa = 10.39VNNIQTSVVTPDD249 pKa = 3.35TSSIWAYY256 pKa = 11.25DD257 pKa = 3.55DD258 pKa = 3.4VTLNGAITVNDD269 pKa = 3.8GLYY272 pKa = 10.47FGQGTNGKK280 pKa = 7.1TLAVDD285 pKa = 3.92GDD287 pKa = 4.11VNLAAEE293 pKa = 4.74DD294 pKa = 3.87LQISGMSRR302 pKa = 11.84AANVVINGVVSGNHH316 pKa = 6.39SIFVYY321 pKa = 10.73SSTLTLAGTNTYY333 pKa = 9.57TGATYY338 pKa = 8.2ATSNNATIEE347 pKa = 4.17VTNQSGLGTSGAGTQIGDD365 pKa = 3.62GDD367 pKa = 3.92SLEE370 pKa = 4.76FNFASAQTVSEE381 pKa = 4.16PLYY384 pKa = 11.22VMGTGVSGTGAVVQTGASTTLSGAIALAGDD414 pKa = 3.58TTFGVDD420 pKa = 3.43SNALFSAFMLSGVISGTGNITVTTSPTTVVANSSIQFTGASPNTYY465 pKa = 8.79TGKK468 pKa = 8.82LTVQGVRR475 pKa = 11.84FYY477 pKa = 10.19PSKK480 pKa = 10.13AASVVAVTGDD490 pKa = 3.38MDD492 pKa = 4.11VLASASKK499 pKa = 10.15PSTVEE504 pKa = 3.72TAFSEE509 pKa = 4.82SIADD513 pKa = 3.63TSHH516 pKa = 4.56VHH518 pKa = 5.85LVKK521 pKa = 10.88DD522 pKa = 4.34GSNKK526 pKa = 9.89ASLSIGSSATEE537 pKa = 3.96TIGYY541 pKa = 6.74VTGDD545 pKa = 3.56GEE547 pKa = 4.88LSVGANGAAVVLTSNADD564 pKa = 3.37YY565 pKa = 9.86TFSGTVSKK573 pKa = 10.5FSNFPAGDD581 pKa = 3.56SFVVKK586 pKa = 10.32KK587 pKa = 9.82GTGTATLTGGLDD599 pKa = 3.18TSYY602 pKa = 10.72IAGVAPVFGAQEE614 pKa = 3.97GSLILNGVFTEE625 pKa = 4.78AGTQVLSGATLKK637 pKa = 10.25GTGTVGAVTVDD648 pKa = 2.85NGGAVSVGNSPGCMTFGSLTLNSGSTYY675 pKa = 7.16TQEE678 pKa = 3.95ISGTTACTQYY688 pKa = 11.4DD689 pKa = 3.67QATVTGAAVLGNATLATTLSTNPADD714 pKa = 3.54GTVFTILTAASVSGTFNGLPDD735 pKa = 4.0GATVNVNGVTLRR747 pKa = 11.84INYY750 pKa = 7.47TATSVTLTKK759 pKa = 10.73VGGNLAPTGSSTAIYY774 pKa = 10.08GLLGLCMVLLSTLALRR790 pKa = 11.84RR791 pKa = 11.84YY792 pKa = 9.74RR793 pKa = 11.84FNNHH797 pKa = 4.62TNN799 pKa = 3.14
Molecular weight: 79.89 kDa
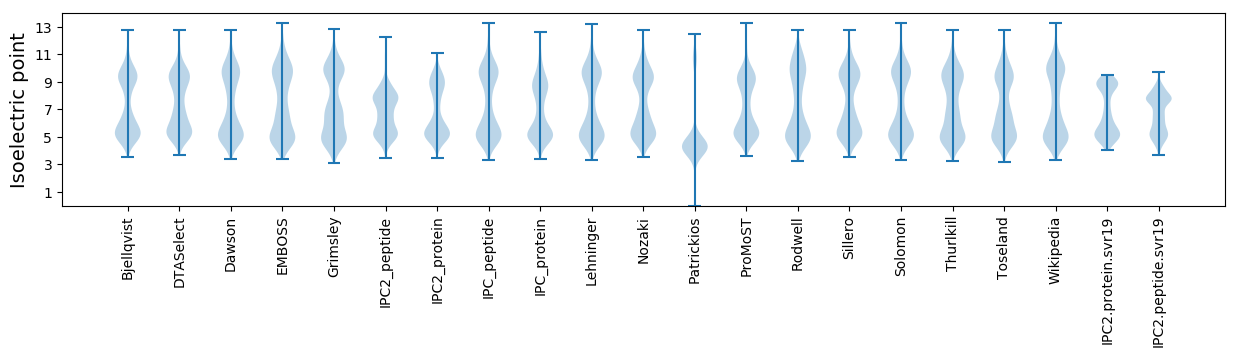
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R4PYS9|R4PYS9_9BACT Uncharacterized protein OS=Candidatus Saccharimonas aalborgensis OX=1332188 GN=L336_0686 PE=4 SV=1
MM1 pKa = 7.84PKK3 pKa = 10.55LKK5 pKa = 8.66THH7 pKa = 6.88KK8 pKa = 9.69GTAKK12 pKa = 10.41RR13 pKa = 11.84IKK15 pKa = 9.53LTSTGKK21 pKa = 8.2LTRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84AFGNHH31 pKa = 5.04MLSKK35 pKa = 10.15KK36 pKa = 10.01SKK38 pKa = 9.02SRR40 pKa = 11.84KK41 pKa = 7.48RR42 pKa = 11.84TINTSATVSGSMAKK56 pKa = 10.22NIKK59 pKa = 9.86RR60 pKa = 11.84VLGVV64 pKa = 3.17
MM1 pKa = 7.84PKK3 pKa = 10.55LKK5 pKa = 8.66THH7 pKa = 6.88KK8 pKa = 9.69GTAKK12 pKa = 10.41RR13 pKa = 11.84IKK15 pKa = 9.53LTSTGKK21 pKa = 8.2LTRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84AFGNHH31 pKa = 5.04MLSKK35 pKa = 10.15KK36 pKa = 10.01SKK38 pKa = 9.02SRR40 pKa = 11.84KK41 pKa = 7.48RR42 pKa = 11.84TINTSATVSGSMAKK56 pKa = 10.22NIKK59 pKa = 9.86RR60 pKa = 11.84VLGVV64 pKa = 3.17
Molecular weight: 7.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
304259 |
27 |
1885 |
294.5 |
32.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.993 ± 0.084 | 0.685 ± 0.023 |
5.701 ± 0.061 | 5.715 ± 0.09 |
3.541 ± 0.051 | 7.13 ± 0.103 |
2.235 ± 0.038 | 6.498 ± 0.063 |
5.135 ± 0.077 | 9.269 ± 0.084 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.347 ± 0.037 | 3.686 ± 0.063 |
4.114 ± 0.055 | 3.825 ± 0.041 |
5.607 ± 0.067 | 6.715 ± 0.081 |
6.621 ± 0.096 | 7.605 ± 0.061 |
1.133 ± 0.029 | 3.444 ± 0.048 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
