
Salinihabitans flavidus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Salinihabitans
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
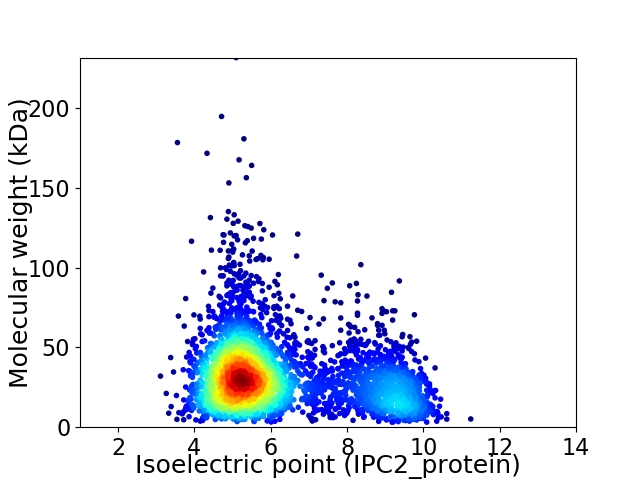
Virtual 2D-PAGE plot for 3934 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H8T3X1|A0A1H8T3X1_9RHOB Uncharacterized protein OS=Salinihabitans flavidus OX=569882 GN=SAMN04490248_11421 PE=4 SV=1
MM1 pKa = 7.89ILIALVAGLLVGGSYY16 pKa = 11.19LLYY19 pKa = 10.39LQFDD23 pKa = 4.45RR24 pKa = 11.84GCADD28 pKa = 3.61ADD30 pKa = 3.93CFVEE34 pKa = 5.59AVQACEE40 pKa = 4.03PEE42 pKa = 4.32TFRR45 pKa = 11.84TQSPAGTGAEE55 pKa = 3.79GVYY58 pKa = 10.01RR59 pKa = 11.84VVGPAGPACEE69 pKa = 3.94ISFEE73 pKa = 4.21YY74 pKa = 10.74SEE76 pKa = 4.6NPNPTFVDD84 pKa = 3.64KK85 pKa = 10.79PVTFVLSPDD94 pKa = 3.47EE95 pKa = 4.47ASEE98 pKa = 4.11DD99 pKa = 3.94TVLGALEE106 pKa = 4.15ACMTGRR112 pKa = 11.84TGPYY116 pKa = 9.06QCGGPLFDD124 pKa = 4.78QLAGSAPAYY133 pKa = 10.44ASLEE137 pKa = 4.12YY138 pKa = 10.91GGDD141 pKa = 3.76GPLPCGDD148 pKa = 3.75PVEE151 pKa = 4.86GAGEE155 pKa = 4.1PLYY158 pKa = 10.21PMPKK162 pKa = 9.47DD163 pKa = 3.83GQWGYY168 pKa = 7.36VTRR171 pKa = 11.84DD172 pKa = 3.84GEE174 pKa = 4.39WQIQPQWDD182 pKa = 3.47RR183 pKa = 11.84AMDD186 pKa = 3.68FHH188 pKa = 7.39EE189 pKa = 4.82GRR191 pKa = 11.84AIVGGKK197 pKa = 8.37NAWGIIDD204 pKa = 4.17RR205 pKa = 11.84DD206 pKa = 3.86GEE208 pKa = 4.18EE209 pKa = 4.3IIAPEE214 pKa = 4.2HH215 pKa = 6.42ASDD218 pKa = 5.1VDD220 pKa = 3.72GQPPFTPYY228 pKa = 10.61SEE230 pKa = 4.45GCTVANIFTDD240 pKa = 4.04TSQPAFFLDD249 pKa = 3.98RR250 pKa = 11.84DD251 pKa = 3.81GKK253 pKa = 10.93AYY255 pKa = 9.5WRR257 pKa = 11.84DD258 pKa = 3.46EE259 pKa = 4.16RR260 pKa = 11.84PEE262 pKa = 3.84ALAALEE268 pKa = 4.34IKK270 pKa = 10.85NFGNFSEE277 pKa = 4.41GLAWFWDD284 pKa = 3.47GDD286 pKa = 4.18IIEE289 pKa = 5.13PSYY292 pKa = 11.47GWIDD296 pKa = 3.35ASGSVAIEE304 pKa = 4.09PEE306 pKa = 4.23FVDD309 pKa = 5.0AGDD312 pKa = 3.83FSGGLAPASSRR323 pKa = 11.84EE324 pKa = 4.03GQAGFISPDD333 pKa = 2.92GRR335 pKa = 11.84LVLPRR340 pKa = 11.84KK341 pKa = 6.46WTLHH345 pKa = 4.93NAQPFSEE352 pKa = 4.6GLAQVWTDD360 pKa = 3.57AFDD363 pKa = 3.28IAYY366 pKa = 9.61MNDD369 pKa = 3.06SDD371 pKa = 5.22FAFHH375 pKa = 7.74DD376 pKa = 3.9VTFPAADD383 pKa = 3.47GSDD386 pKa = 3.45PVRR389 pKa = 11.84GQIEE393 pKa = 4.17AAGPFHH399 pKa = 7.74DD400 pKa = 4.62GLAPVVAVYY409 pKa = 10.84DD410 pKa = 3.8GTTEE414 pKa = 4.02FAYY417 pKa = 10.09MNQQGEE423 pKa = 4.6VAFVPDD429 pKa = 4.17RR430 pKa = 11.84LDD432 pKa = 5.22GIALCDD438 pKa = 3.28ARR440 pKa = 11.84LRR442 pKa = 11.84PEE444 pKa = 3.88FHH446 pKa = 6.67NGLVRR451 pKa = 11.84LVVADD456 pKa = 4.7DD457 pKa = 4.56GEE459 pKa = 4.78SCGDD463 pKa = 3.37EE464 pKa = 4.42SYY466 pKa = 11.71LDD468 pKa = 3.68GAPAYY473 pKa = 9.89DD474 pKa = 3.66AAHH477 pKa = 6.23YY478 pKa = 10.27LYY480 pKa = 10.66LDD482 pKa = 3.5TQGRR486 pKa = 11.84VAMQQVKK493 pKa = 10.21
MM1 pKa = 7.89ILIALVAGLLVGGSYY16 pKa = 11.19LLYY19 pKa = 10.39LQFDD23 pKa = 4.45RR24 pKa = 11.84GCADD28 pKa = 3.61ADD30 pKa = 3.93CFVEE34 pKa = 5.59AVQACEE40 pKa = 4.03PEE42 pKa = 4.32TFRR45 pKa = 11.84TQSPAGTGAEE55 pKa = 3.79GVYY58 pKa = 10.01RR59 pKa = 11.84VVGPAGPACEE69 pKa = 3.94ISFEE73 pKa = 4.21YY74 pKa = 10.74SEE76 pKa = 4.6NPNPTFVDD84 pKa = 3.64KK85 pKa = 10.79PVTFVLSPDD94 pKa = 3.47EE95 pKa = 4.47ASEE98 pKa = 4.11DD99 pKa = 3.94TVLGALEE106 pKa = 4.15ACMTGRR112 pKa = 11.84TGPYY116 pKa = 9.06QCGGPLFDD124 pKa = 4.78QLAGSAPAYY133 pKa = 10.44ASLEE137 pKa = 4.12YY138 pKa = 10.91GGDD141 pKa = 3.76GPLPCGDD148 pKa = 3.75PVEE151 pKa = 4.86GAGEE155 pKa = 4.1PLYY158 pKa = 10.21PMPKK162 pKa = 9.47DD163 pKa = 3.83GQWGYY168 pKa = 7.36VTRR171 pKa = 11.84DD172 pKa = 3.84GEE174 pKa = 4.39WQIQPQWDD182 pKa = 3.47RR183 pKa = 11.84AMDD186 pKa = 3.68FHH188 pKa = 7.39EE189 pKa = 4.82GRR191 pKa = 11.84AIVGGKK197 pKa = 8.37NAWGIIDD204 pKa = 4.17RR205 pKa = 11.84DD206 pKa = 3.86GEE208 pKa = 4.18EE209 pKa = 4.3IIAPEE214 pKa = 4.2HH215 pKa = 6.42ASDD218 pKa = 5.1VDD220 pKa = 3.72GQPPFTPYY228 pKa = 10.61SEE230 pKa = 4.45GCTVANIFTDD240 pKa = 4.04TSQPAFFLDD249 pKa = 3.98RR250 pKa = 11.84DD251 pKa = 3.81GKK253 pKa = 10.93AYY255 pKa = 9.5WRR257 pKa = 11.84DD258 pKa = 3.46EE259 pKa = 4.16RR260 pKa = 11.84PEE262 pKa = 3.84ALAALEE268 pKa = 4.34IKK270 pKa = 10.85NFGNFSEE277 pKa = 4.41GLAWFWDD284 pKa = 3.47GDD286 pKa = 4.18IIEE289 pKa = 5.13PSYY292 pKa = 11.47GWIDD296 pKa = 3.35ASGSVAIEE304 pKa = 4.09PEE306 pKa = 4.23FVDD309 pKa = 5.0AGDD312 pKa = 3.83FSGGLAPASSRR323 pKa = 11.84EE324 pKa = 4.03GQAGFISPDD333 pKa = 2.92GRR335 pKa = 11.84LVLPRR340 pKa = 11.84KK341 pKa = 6.46WTLHH345 pKa = 4.93NAQPFSEE352 pKa = 4.6GLAQVWTDD360 pKa = 3.57AFDD363 pKa = 3.28IAYY366 pKa = 9.61MNDD369 pKa = 3.06SDD371 pKa = 5.22FAFHH375 pKa = 7.74DD376 pKa = 3.9VTFPAADD383 pKa = 3.47GSDD386 pKa = 3.45PVRR389 pKa = 11.84GQIEE393 pKa = 4.17AAGPFHH399 pKa = 7.74DD400 pKa = 4.62GLAPVVAVYY409 pKa = 10.84DD410 pKa = 3.8GTTEE414 pKa = 4.02FAYY417 pKa = 10.09MNQQGEE423 pKa = 4.6VAFVPDD429 pKa = 4.17RR430 pKa = 11.84LDD432 pKa = 5.22GIALCDD438 pKa = 3.28ARR440 pKa = 11.84LRR442 pKa = 11.84PEE444 pKa = 3.88FHH446 pKa = 6.67NGLVRR451 pKa = 11.84LVVADD456 pKa = 4.7DD457 pKa = 4.56GEE459 pKa = 4.78SCGDD463 pKa = 3.37EE464 pKa = 4.42SYY466 pKa = 11.71LDD468 pKa = 3.68GAPAYY473 pKa = 9.89DD474 pKa = 3.66AAHH477 pKa = 6.23YY478 pKa = 10.27LYY480 pKa = 10.66LDD482 pKa = 3.5TQGRR486 pKa = 11.84VAMQQVKK493 pKa = 10.21
Molecular weight: 53.06 kDa
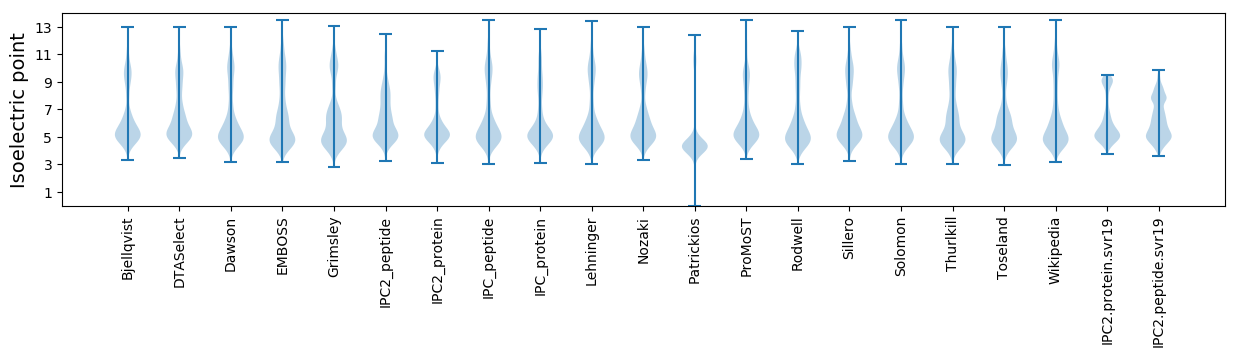
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H8SH60|A0A1H8SH60_9RHOB Riboflavin biosynthesis protein OS=Salinihabitans flavidus OX=569882 GN=SAMN04490248_11224 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1213260 |
28 |
2144 |
308.4 |
33.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.146 ± 0.052 | 0.896 ± 0.011 |
6.053 ± 0.034 | 6.192 ± 0.038 |
3.686 ± 0.025 | 8.86 ± 0.044 |
2.149 ± 0.018 | 5.122 ± 0.03 |
2.818 ± 0.032 | 10.023 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.816 ± 0.018 | 2.437 ± 0.022 |
5.15 ± 0.026 | 3.03 ± 0.021 |
7.31 ± 0.043 | 5.043 ± 0.026 |
5.4 ± 0.025 | 7.291 ± 0.035 |
1.412 ± 0.017 | 2.165 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
