
Amnibacterium flavum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Amnibacterium
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
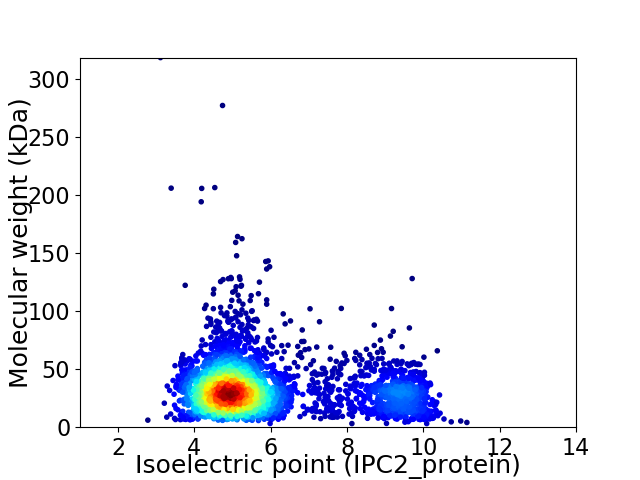
Virtual 2D-PAGE plot for 3308 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V1HSZ4|A0A2V1HSZ4_9MICO Glucose-6-phosphate dehydrogenase OS=Amnibacterium flavum OX=2173173 GN=DDQ50_10025 PE=4 SV=1
MM1 pKa = 8.03DD2 pKa = 5.1PCILPSTGFDD12 pKa = 3.37PSLLLGLTAVAILGGGLSVWAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84SKK39 pKa = 10.86RR40 pKa = 11.84SGVALAVPFLVLLLLAGSQVTPPAASAAVACPPAPAGAPATGGGTTAPVTFEE92 pKa = 4.75PGSAGIGDD100 pKa = 4.59PYY102 pKa = 11.24YY103 pKa = 10.22PLDD106 pKa = 3.92GNGGYY111 pKa = 10.15DD112 pKa = 3.46VQNYY116 pKa = 10.19DD117 pKa = 4.53LDD119 pKa = 4.18LAYY122 pKa = 10.6DD123 pKa = 3.83PATDD127 pKa = 3.75VLAGTATIDD136 pKa = 4.58LVATQNLSAFNLDD149 pKa = 3.11FDD151 pKa = 4.18TRR153 pKa = 11.84DD154 pKa = 3.14AAGADD159 pKa = 3.84SIVIGDD165 pKa = 3.63VRR167 pKa = 11.84VDD169 pKa = 3.56GASTAFSLASTQISLVTAQPVGDD192 pKa = 4.4GSPDD196 pKa = 3.21AVAEE200 pKa = 4.26PPRR203 pKa = 11.84TEE205 pKa = 4.23LTVMPATGILSGTEE219 pKa = 3.59LSVEE223 pKa = 3.62VDD225 pKa = 3.49YY226 pKa = 11.72AGVPITIDD234 pKa = 3.31DD235 pKa = 3.93AFGLGGVLHH244 pKa = 6.68TPDD247 pKa = 2.92GMVPVGQPRR256 pKa = 11.84AAAVWFPSNDD266 pKa = 3.2HH267 pKa = 6.81PADD270 pKa = 3.67KK271 pKa = 10.26ATLTMSMTVPIGLDD285 pKa = 3.28VIGNGVLASTVDD297 pKa = 3.58NGPTSTWTYY306 pKa = 10.86VMDD309 pKa = 4.54RR310 pKa = 11.84PMATYY315 pKa = 10.4LATAVVGDD323 pKa = 4.29YY324 pKa = 10.81DD325 pKa = 4.0VQTTTVDD332 pKa = 3.0GVTYY336 pKa = 10.21RR337 pKa = 11.84DD338 pKa = 3.45AVAQSLYY345 pKa = 10.96SQGTLGADD353 pKa = 2.89ARR355 pKa = 11.84TALDD359 pKa = 4.2LNPDD363 pKa = 4.07VIAYY367 pKa = 9.55LSSIYY372 pKa = 10.49GPYY375 pKa = 10.28PFTEE379 pKa = 4.17SGGIAVDD386 pKa = 5.12LPDD389 pKa = 5.07LGYY392 pKa = 11.25ALEE395 pKa = 4.36NQTRR399 pKa = 11.84PIYY402 pKa = 10.75GFIDD406 pKa = 4.01EE407 pKa = 4.51PTVVHH412 pKa = 6.5EE413 pKa = 5.68LAHH416 pKa = 5.23QWYY419 pKa = 9.07GDD421 pKa = 3.62DD422 pKa = 3.61VAIEE426 pKa = 4.08RR427 pKa = 11.84WSDD430 pKa = 2.35IWLNEE435 pKa = 3.39AFATYY440 pKa = 10.27SEE442 pKa = 4.41WLWSEE447 pKa = 3.98DD448 pKa = 3.39HH449 pKa = 7.06GGDD452 pKa = 3.19TAQQIFDD459 pKa = 3.94DD460 pKa = 4.87TYY462 pKa = 11.72SLDD465 pKa = 3.51ASDD468 pKa = 4.88PFWSVEE474 pKa = 4.02VADD477 pKa = 4.81PGAPRR482 pKa = 11.84LFDD485 pKa = 3.21NATYY489 pKa = 10.82YY490 pKa = 10.43RR491 pKa = 11.84GAMTLHH497 pKa = 7.38ALRR500 pKa = 11.84LEE502 pKa = 4.39VGDD505 pKa = 3.87PVFFGILRR513 pKa = 11.84GWAADD518 pKa = 3.55NAGGNVSTAQFEE530 pKa = 4.87AYY532 pKa = 10.09ASAAAGTDD540 pKa = 3.21LSGFFDD546 pKa = 3.66TWIHH550 pKa = 6.61SSVKK554 pKa = 9.95PP555 pKa = 3.61
MM1 pKa = 8.03DD2 pKa = 5.1PCILPSTGFDD12 pKa = 3.37PSLLLGLTAVAILGGGLSVWAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84SKK39 pKa = 10.86RR40 pKa = 11.84SGVALAVPFLVLLLLAGSQVTPPAASAAVACPPAPAGAPATGGGTTAPVTFEE92 pKa = 4.75PGSAGIGDD100 pKa = 4.59PYY102 pKa = 11.24YY103 pKa = 10.22PLDD106 pKa = 3.92GNGGYY111 pKa = 10.15DD112 pKa = 3.46VQNYY116 pKa = 10.19DD117 pKa = 4.53LDD119 pKa = 4.18LAYY122 pKa = 10.6DD123 pKa = 3.83PATDD127 pKa = 3.75VLAGTATIDD136 pKa = 4.58LVATQNLSAFNLDD149 pKa = 3.11FDD151 pKa = 4.18TRR153 pKa = 11.84DD154 pKa = 3.14AAGADD159 pKa = 3.84SIVIGDD165 pKa = 3.63VRR167 pKa = 11.84VDD169 pKa = 3.56GASTAFSLASTQISLVTAQPVGDD192 pKa = 4.4GSPDD196 pKa = 3.21AVAEE200 pKa = 4.26PPRR203 pKa = 11.84TEE205 pKa = 4.23LTVMPATGILSGTEE219 pKa = 3.59LSVEE223 pKa = 3.62VDD225 pKa = 3.49YY226 pKa = 11.72AGVPITIDD234 pKa = 3.31DD235 pKa = 3.93AFGLGGVLHH244 pKa = 6.68TPDD247 pKa = 2.92GMVPVGQPRR256 pKa = 11.84AAAVWFPSNDD266 pKa = 3.2HH267 pKa = 6.81PADD270 pKa = 3.67KK271 pKa = 10.26ATLTMSMTVPIGLDD285 pKa = 3.28VIGNGVLASTVDD297 pKa = 3.58NGPTSTWTYY306 pKa = 10.86VMDD309 pKa = 4.54RR310 pKa = 11.84PMATYY315 pKa = 10.4LATAVVGDD323 pKa = 4.29YY324 pKa = 10.81DD325 pKa = 4.0VQTTTVDD332 pKa = 3.0GVTYY336 pKa = 10.21RR337 pKa = 11.84DD338 pKa = 3.45AVAQSLYY345 pKa = 10.96SQGTLGADD353 pKa = 2.89ARR355 pKa = 11.84TALDD359 pKa = 4.2LNPDD363 pKa = 4.07VIAYY367 pKa = 9.55LSSIYY372 pKa = 10.49GPYY375 pKa = 10.28PFTEE379 pKa = 4.17SGGIAVDD386 pKa = 5.12LPDD389 pKa = 5.07LGYY392 pKa = 11.25ALEE395 pKa = 4.36NQTRR399 pKa = 11.84PIYY402 pKa = 10.75GFIDD406 pKa = 4.01EE407 pKa = 4.51PTVVHH412 pKa = 6.5EE413 pKa = 5.68LAHH416 pKa = 5.23QWYY419 pKa = 9.07GDD421 pKa = 3.62DD422 pKa = 3.61VAIEE426 pKa = 4.08RR427 pKa = 11.84WSDD430 pKa = 2.35IWLNEE435 pKa = 3.39AFATYY440 pKa = 10.27SEE442 pKa = 4.41WLWSEE447 pKa = 3.98DD448 pKa = 3.39HH449 pKa = 7.06GGDD452 pKa = 3.19TAQQIFDD459 pKa = 3.94DD460 pKa = 4.87TYY462 pKa = 11.72SLDD465 pKa = 3.51ASDD468 pKa = 4.88PFWSVEE474 pKa = 4.02VADD477 pKa = 4.81PGAPRR482 pKa = 11.84LFDD485 pKa = 3.21NATYY489 pKa = 10.82YY490 pKa = 10.43RR491 pKa = 11.84GAMTLHH497 pKa = 7.38ALRR500 pKa = 11.84LEE502 pKa = 4.39VGDD505 pKa = 3.87PVFFGILRR513 pKa = 11.84GWAADD518 pKa = 3.55NAGGNVSTAQFEE530 pKa = 4.87AYY532 pKa = 10.09ASAAAGTDD540 pKa = 3.21LSGFFDD546 pKa = 3.66TWIHH550 pKa = 6.61SSVKK554 pKa = 9.95PP555 pKa = 3.61
Molecular weight: 57.97 kDa
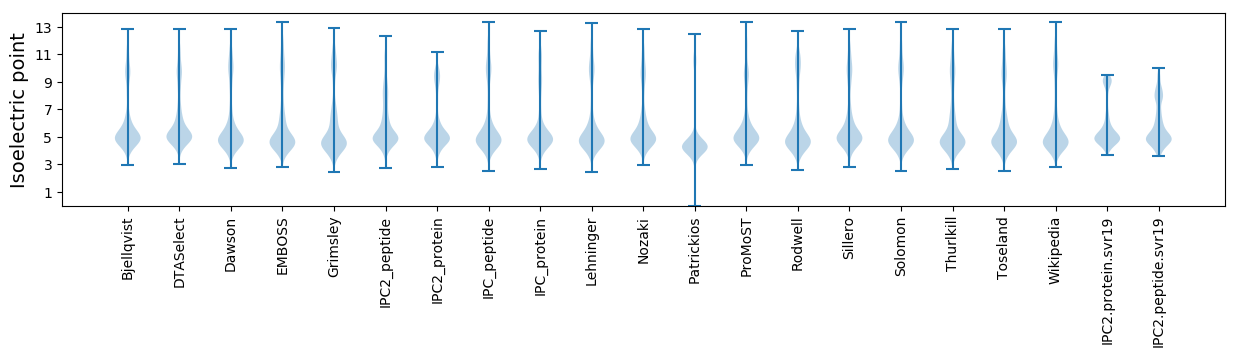
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V1HWS1|A0A2V1HWS1_9MICO GNAT family N-acetyltransferase OS=Amnibacterium flavum OX=2173173 GN=DDQ50_04145 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1063717 |
28 |
3212 |
321.6 |
34.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.001 ± 0.061 | 0.457 ± 0.011 |
6.368 ± 0.052 | 5.598 ± 0.045 |
3.169 ± 0.027 | 9.154 ± 0.042 |
1.787 ± 0.021 | 4.824 ± 0.032 |
1.906 ± 0.029 | 10.155 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.662 ± 0.015 | 1.999 ± 0.031 |
5.38 ± 0.036 | 2.635 ± 0.022 |
7.143 ± 0.055 | 6.251 ± 0.036 |
6.111 ± 0.045 | 8.948 ± 0.036 |
1.427 ± 0.015 | 2.025 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
