
Halorubrum sp. AJ67
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Halorubrum; unclassified Halorubrum
Average proteome isoelectric point is 5.65
Get precalculated fractions of proteins
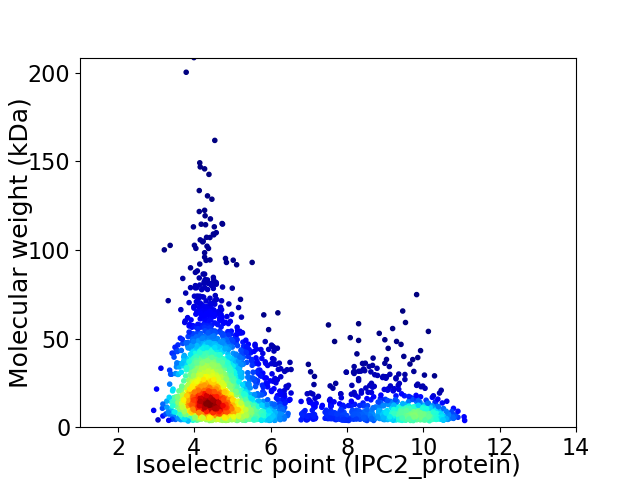
Virtual 2D-PAGE plot for 3364 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
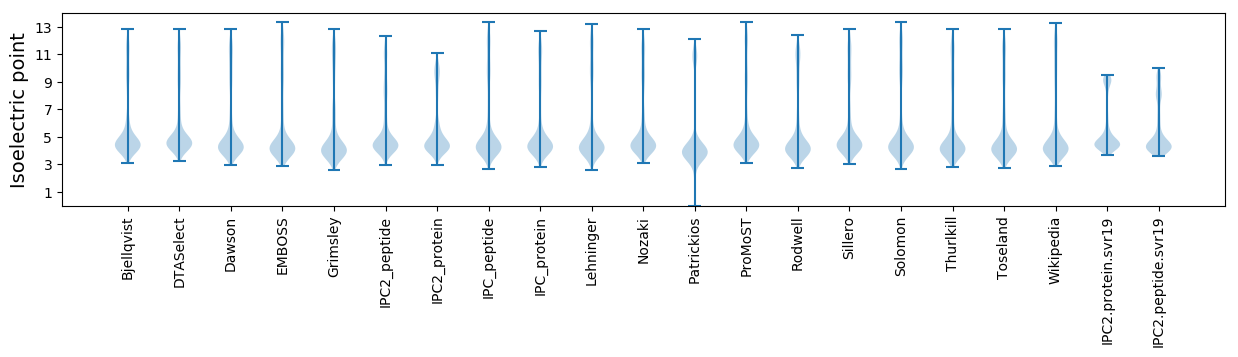
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V6DWC7|V6DWC7_9EURY Isoform of V6DXC1 Nucleotidyltransferase OS=Halorubrum sp. AJ67 OX=1173487 GN=C483_00770 PE=4 SV=1
MM1 pKa = 7.35SFDD4 pKa = 4.2TIFVPAFGSDD14 pKa = 2.8ASRR17 pKa = 11.84ILEE20 pKa = 4.16IDD22 pKa = 3.55NEE24 pKa = 4.54TGDD27 pKa = 3.73TVAFHH32 pKa = 6.6SVSDD36 pKa = 3.83SVGGIDD42 pKa = 5.22RR43 pKa = 11.84DD44 pKa = 3.46ADD46 pKa = 3.74GNVYY50 pKa = 10.49AIVGQYY56 pKa = 9.49LKK58 pKa = 10.6KK59 pKa = 10.69YY60 pKa = 9.4SPDD63 pKa = 3.22MNLIWTSGEE72 pKa = 4.34TYY74 pKa = 8.7STTVVEE80 pKa = 4.79VADD83 pKa = 3.74NGNAIFVGGGSRR95 pKa = 11.84IIRR98 pKa = 11.84LDD100 pKa = 3.48PDD102 pKa = 3.99GSIVWNSYY110 pKa = 8.13DD111 pKa = 3.82TNIGCLDD118 pKa = 3.47MSYY121 pKa = 11.31DD122 pKa = 3.7GGDD125 pKa = 3.53YY126 pKa = 10.77LYY128 pKa = 11.16VQAADD133 pKa = 3.7DD134 pKa = 4.21SEE136 pKa = 3.84DD137 pKa = 3.36RR138 pKa = 11.84RR139 pKa = 11.84YY140 pKa = 11.06GFANGNDD147 pKa = 3.66SAEE150 pKa = 4.14EE151 pKa = 3.72LVGYY155 pKa = 10.56NDD157 pKa = 4.18DD158 pKa = 3.56SSGILHH164 pKa = 6.86HH165 pKa = 6.72NGSLYY170 pKa = 9.91STEE173 pKa = 4.17LTTSNLWRR181 pKa = 11.84SDD183 pKa = 3.31TFVEE187 pKa = 5.22GSTINDD193 pKa = 3.42GNFTVSNYY201 pKa = 9.67TNLDD205 pKa = 3.11MDD207 pKa = 4.15RR208 pKa = 11.84GGLYY212 pKa = 9.57TAGSGGKK219 pKa = 8.69IAHH222 pKa = 7.13LDD224 pKa = 3.62FDD226 pKa = 6.1GNTQWEE232 pKa = 4.46VSISGSPYY240 pKa = 9.6FVGIDD245 pKa = 3.48VSPDD249 pKa = 3.61TLWASSDD256 pKa = 3.15AGLYY260 pKa = 10.46RR261 pKa = 11.84IDD263 pKa = 4.16KK264 pKa = 10.77SDD266 pKa = 4.8GSVLWKK272 pKa = 9.39NTDD275 pKa = 3.12TQFNSTAHH283 pKa = 6.27EE284 pKa = 4.31SLGGFPNLDD293 pKa = 3.61SYY295 pKa = 11.9GGANWEE301 pKa = 4.11VTVQISGTATVDD313 pKa = 3.33GASTSDD319 pKa = 3.56VEE321 pKa = 4.47ILVIDD326 pKa = 4.09EE327 pKa = 4.92DD328 pKa = 4.68SNAFGGRR335 pKa = 11.84TTTDD339 pKa = 2.85SNGNWTVEE347 pKa = 4.18APDD350 pKa = 3.63STLHH354 pKa = 6.66VVAQYY359 pKa = 11.3KK360 pKa = 10.57DD361 pKa = 3.0ADD363 pKa = 3.67GNVFNTEE370 pKa = 3.7SYY372 pKa = 10.54PYY374 pKa = 10.61VNSS377 pKa = 3.6
MM1 pKa = 7.35SFDD4 pKa = 4.2TIFVPAFGSDD14 pKa = 2.8ASRR17 pKa = 11.84ILEE20 pKa = 4.16IDD22 pKa = 3.55NEE24 pKa = 4.54TGDD27 pKa = 3.73TVAFHH32 pKa = 6.6SVSDD36 pKa = 3.83SVGGIDD42 pKa = 5.22RR43 pKa = 11.84DD44 pKa = 3.46ADD46 pKa = 3.74GNVYY50 pKa = 10.49AIVGQYY56 pKa = 9.49LKK58 pKa = 10.6KK59 pKa = 10.69YY60 pKa = 9.4SPDD63 pKa = 3.22MNLIWTSGEE72 pKa = 4.34TYY74 pKa = 8.7STTVVEE80 pKa = 4.79VADD83 pKa = 3.74NGNAIFVGGGSRR95 pKa = 11.84IIRR98 pKa = 11.84LDD100 pKa = 3.48PDD102 pKa = 3.99GSIVWNSYY110 pKa = 8.13DD111 pKa = 3.82TNIGCLDD118 pKa = 3.47MSYY121 pKa = 11.31DD122 pKa = 3.7GGDD125 pKa = 3.53YY126 pKa = 10.77LYY128 pKa = 11.16VQAADD133 pKa = 3.7DD134 pKa = 4.21SEE136 pKa = 3.84DD137 pKa = 3.36RR138 pKa = 11.84RR139 pKa = 11.84YY140 pKa = 11.06GFANGNDD147 pKa = 3.66SAEE150 pKa = 4.14EE151 pKa = 3.72LVGYY155 pKa = 10.56NDD157 pKa = 4.18DD158 pKa = 3.56SSGILHH164 pKa = 6.86HH165 pKa = 6.72NGSLYY170 pKa = 9.91STEE173 pKa = 4.17LTTSNLWRR181 pKa = 11.84SDD183 pKa = 3.31TFVEE187 pKa = 5.22GSTINDD193 pKa = 3.42GNFTVSNYY201 pKa = 9.67TNLDD205 pKa = 3.11MDD207 pKa = 4.15RR208 pKa = 11.84GGLYY212 pKa = 9.57TAGSGGKK219 pKa = 8.69IAHH222 pKa = 7.13LDD224 pKa = 3.62FDD226 pKa = 6.1GNTQWEE232 pKa = 4.46VSISGSPYY240 pKa = 9.6FVGIDD245 pKa = 3.48VSPDD249 pKa = 3.61TLWASSDD256 pKa = 3.15AGLYY260 pKa = 10.46RR261 pKa = 11.84IDD263 pKa = 4.16KK264 pKa = 10.77SDD266 pKa = 4.8GSVLWKK272 pKa = 9.39NTDD275 pKa = 3.12TQFNSTAHH283 pKa = 6.27EE284 pKa = 4.31SLGGFPNLDD293 pKa = 3.61SYY295 pKa = 11.9GGANWEE301 pKa = 4.11VTVQISGTATVDD313 pKa = 3.33GASTSDD319 pKa = 3.56VEE321 pKa = 4.47ILVIDD326 pKa = 4.09EE327 pKa = 4.92DD328 pKa = 4.68SNAFGGRR335 pKa = 11.84TTTDD339 pKa = 2.85SNGNWTVEE347 pKa = 4.18APDD350 pKa = 3.63STLHH354 pKa = 6.66VVAQYY359 pKa = 11.3KK360 pKa = 10.57DD361 pKa = 3.0ADD363 pKa = 3.67GNVFNTEE370 pKa = 3.7SYY372 pKa = 10.54PYY374 pKa = 10.61VNSS377 pKa = 3.6
Molecular weight: 40.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V6DTB7|V6DTB7_9EURY Isoform of V6DQ87 Transposase IS605 OrfB family protein OS=Halorubrum sp. AJ67 OX=1173487 GN=C468_01355 PE=4 SV=1
MM1 pKa = 7.37GVPVVGGKK9 pKa = 7.96AHH11 pKa = 6.86RR12 pKa = 11.84GVGXXXXXXXPSRR25 pKa = 11.84RR26 pKa = 11.84ARR28 pKa = 11.84GRR30 pKa = 11.84SAPTAASRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84LSATT44 pKa = 3.11
MM1 pKa = 7.37GVPVVGGKK9 pKa = 7.96AHH11 pKa = 6.86RR12 pKa = 11.84GVGXXXXXXXPSRR25 pKa = 11.84RR26 pKa = 11.84ARR28 pKa = 11.84GRR30 pKa = 11.84SAPTAASRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84LSATT44 pKa = 3.11
Molecular weight: 3.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
684466 |
37 |
1978 |
203.5 |
22.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.647 ± 0.07 | 0.791 ± 0.016 |
8.322 ± 0.057 | 8.354 ± 0.065 |
3.43 ± 0.035 | 8.139 ± 0.054 |
2.05 ± 0.03 | 4.52 ± 0.037 |
2.176 ± 0.032 | 8.497 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.878 ± 0.024 | 2.698 ± 0.034 |
4.7 ± 0.035 | 2.642 ± 0.029 |
7.009 ± 0.071 | 6.186 ± 0.049 |
6.413 ± 0.042 | 8.519 ± 0.052 |
1.158 ± 0.022 | 2.805 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
