
Pteronotus davyi polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
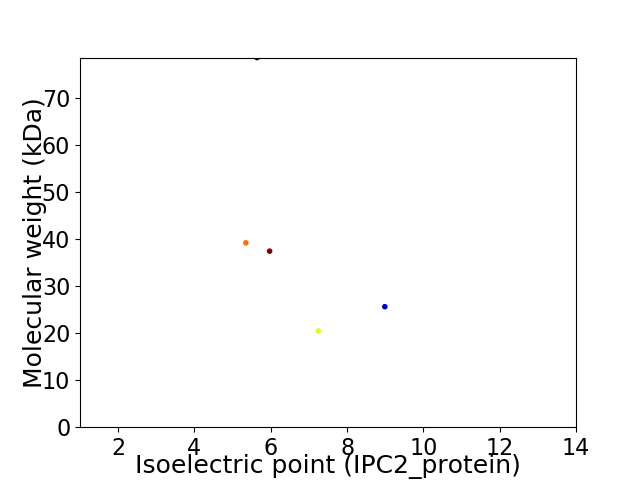
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L0GAK6|L0GAK6_9POLY Minor capsid protein OS=Pteronotus davyi polyomavirus 1 OX=1891773 PE=3 SV=1
MM1 pKa = 7.85APPQKK6 pKa = 10.21RR7 pKa = 11.84SPKK10 pKa = 9.76LPAPAAVPKK19 pKa = 10.55LLIKK23 pKa = 10.78GGIEE27 pKa = 4.01VLDD30 pKa = 4.61LKK32 pKa = 10.08TGPDD36 pKa = 3.99SITQIEE42 pKa = 4.82LFLNPRR48 pKa = 11.84MGQPTTSADD57 pKa = 2.93WVGFSDD63 pKa = 3.81NVTVSASWSADD74 pKa = 3.01SPAANQTPCYY84 pKa = 9.18SCARR88 pKa = 11.84VSLPMINEE96 pKa = 4.21DD97 pKa = 3.41MTSDD101 pKa = 5.06KK102 pKa = 10.99ILMWEE107 pKa = 3.97AVSCKK112 pKa = 9.79TEE114 pKa = 3.96VVGVSSLTNVHH125 pKa = 5.55SFKK128 pKa = 10.99KK129 pKa = 10.03KK130 pKa = 9.17LHH132 pKa = 6.27DD133 pKa = 3.8NEE135 pKa = 5.77GIGLPVEE142 pKa = 4.11GLNYY146 pKa = 10.63HH147 pKa = 6.25MFAVGGEE154 pKa = 3.94PLEE157 pKa = 4.1LQFLVEE163 pKa = 4.13NHH165 pKa = 5.3HH166 pKa = 5.56TVYY169 pKa = 10.74GAGLKK174 pKa = 9.27TIQNPGASAQTLNPALKK191 pKa = 9.2GTLDD195 pKa = 3.48KK196 pKa = 11.46DD197 pKa = 3.42GAYY200 pKa = 9.49PIEE203 pKa = 4.63AWCPDD208 pKa = 3.35PSKK211 pKa = 11.45NEE213 pKa = 3.52NTRR216 pKa = 11.84YY217 pKa = 9.48FGSYY221 pKa = 9.33TGGQNTPPVLQFTNTVTTILLDD243 pKa = 3.88EE244 pKa = 5.0NGVGPLCKK252 pKa = 10.12GDD254 pKa = 4.39GLYY257 pKa = 10.67LSCADD262 pKa = 2.71IVGFHH267 pKa = 5.61TQAGNNKK274 pKa = 8.22MKK276 pKa = 10.48FRR278 pKa = 11.84GLPRR282 pKa = 11.84YY283 pKa = 9.98FNVTLRR289 pKa = 11.84KK290 pKa = 9.25RR291 pKa = 11.84VVKK294 pKa = 10.6NPYY297 pKa = 9.06PVSSLLNSLFTKK309 pKa = 9.75LIPKK313 pKa = 9.34MDD315 pKa = 4.33GQPMSGDD322 pKa = 3.06NGQVEE327 pKa = 4.68EE328 pKa = 4.15VRR330 pKa = 11.84VYY332 pKa = 10.83EE333 pKa = 4.28GTEE336 pKa = 4.02EE337 pKa = 4.39VPGDD341 pKa = 3.93PDD343 pKa = 3.24MVRR346 pKa = 11.84YY347 pKa = 8.68IDD349 pKa = 5.08KK350 pKa = 9.77YY351 pKa = 9.33GQMEE355 pKa = 4.48TEE357 pKa = 4.6VPQQ360 pKa = 3.9
MM1 pKa = 7.85APPQKK6 pKa = 10.21RR7 pKa = 11.84SPKK10 pKa = 9.76LPAPAAVPKK19 pKa = 10.55LLIKK23 pKa = 10.78GGIEE27 pKa = 4.01VLDD30 pKa = 4.61LKK32 pKa = 10.08TGPDD36 pKa = 3.99SITQIEE42 pKa = 4.82LFLNPRR48 pKa = 11.84MGQPTTSADD57 pKa = 2.93WVGFSDD63 pKa = 3.81NVTVSASWSADD74 pKa = 3.01SPAANQTPCYY84 pKa = 9.18SCARR88 pKa = 11.84VSLPMINEE96 pKa = 4.21DD97 pKa = 3.41MTSDD101 pKa = 5.06KK102 pKa = 10.99ILMWEE107 pKa = 3.97AVSCKK112 pKa = 9.79TEE114 pKa = 3.96VVGVSSLTNVHH125 pKa = 5.55SFKK128 pKa = 10.99KK129 pKa = 10.03KK130 pKa = 9.17LHH132 pKa = 6.27DD133 pKa = 3.8NEE135 pKa = 5.77GIGLPVEE142 pKa = 4.11GLNYY146 pKa = 10.63HH147 pKa = 6.25MFAVGGEE154 pKa = 3.94PLEE157 pKa = 4.1LQFLVEE163 pKa = 4.13NHH165 pKa = 5.3HH166 pKa = 5.56TVYY169 pKa = 10.74GAGLKK174 pKa = 9.27TIQNPGASAQTLNPALKK191 pKa = 9.2GTLDD195 pKa = 3.48KK196 pKa = 11.46DD197 pKa = 3.42GAYY200 pKa = 9.49PIEE203 pKa = 4.63AWCPDD208 pKa = 3.35PSKK211 pKa = 11.45NEE213 pKa = 3.52NTRR216 pKa = 11.84YY217 pKa = 9.48FGSYY221 pKa = 9.33TGGQNTPPVLQFTNTVTTILLDD243 pKa = 3.88EE244 pKa = 5.0NGVGPLCKK252 pKa = 10.12GDD254 pKa = 4.39GLYY257 pKa = 10.67LSCADD262 pKa = 2.71IVGFHH267 pKa = 5.61TQAGNNKK274 pKa = 8.22MKK276 pKa = 10.48FRR278 pKa = 11.84GLPRR282 pKa = 11.84YY283 pKa = 9.98FNVTLRR289 pKa = 11.84KK290 pKa = 9.25RR291 pKa = 11.84VVKK294 pKa = 10.6NPYY297 pKa = 9.06PVSSLLNSLFTKK309 pKa = 9.75LIPKK313 pKa = 9.34MDD315 pKa = 4.33GQPMSGDD322 pKa = 3.06NGQVEE327 pKa = 4.68EE328 pKa = 4.15VRR330 pKa = 11.84VYY332 pKa = 10.83EE333 pKa = 4.28GTEE336 pKa = 4.02EE337 pKa = 4.39VPGDD341 pKa = 3.93PDD343 pKa = 3.24MVRR346 pKa = 11.84YY347 pKa = 8.68IDD349 pKa = 5.08KK350 pKa = 9.77YY351 pKa = 9.33GQMEE355 pKa = 4.48TEE357 pKa = 4.6VPQQ360 pKa = 3.9
Molecular weight: 39.22 kDa
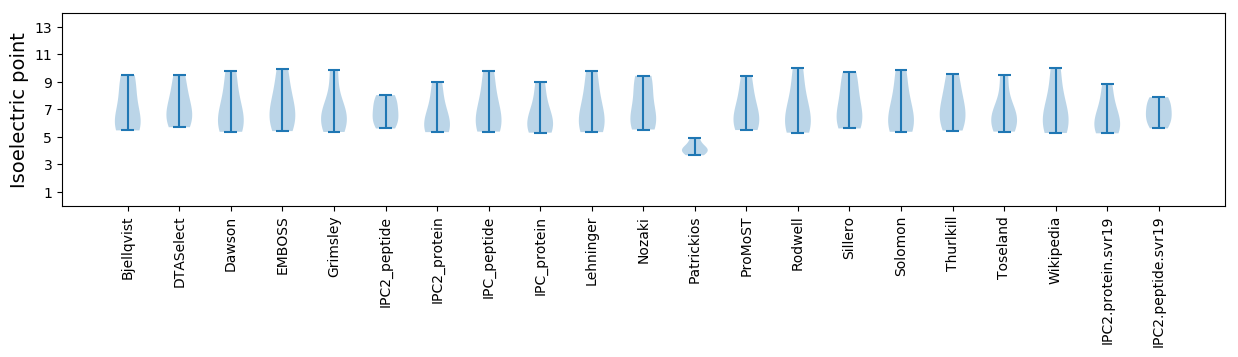
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0GCI1|L0GCI1_9POLY Minor capsid protein OS=Pteronotus davyi polyomavirus 1 OX=1891773 PE=3 SV=1
MM1 pKa = 7.49ALVPYY6 pKa = 9.8MPDD9 pKa = 3.09PDD11 pKa = 3.46IFFPGVSTFARR22 pKa = 11.84YY23 pKa = 9.82ANYY26 pKa = 10.46LDD28 pKa = 4.28PFHH31 pKa = 7.1WAPDD35 pKa = 3.59LYY37 pKa = 10.54HH38 pKa = 6.53QVSRR42 pKa = 11.84WIWEE46 pKa = 4.19SLMHH50 pKa = 6.47EE51 pKa = 4.53GRR53 pKa = 11.84RR54 pKa = 11.84QIGYY58 pKa = 10.03AGQEE62 pKa = 4.04LVRR65 pKa = 11.84YY66 pKa = 7.48TSTTVQDD73 pKa = 3.22TLARR77 pKa = 11.84FFEE80 pKa = 4.17NARR83 pKa = 11.84WAVSHH88 pKa = 6.96FSTNIYY94 pKa = 9.89TSLHH98 pKa = 6.17DD99 pKa = 4.42YY100 pKa = 10.5YY101 pKa = 11.3RR102 pKa = 11.84EE103 pKa = 3.99LPKK106 pKa = 10.7LKK108 pKa = 10.23PEE110 pKa = 4.03QARR113 pKa = 11.84QLSRR117 pKa = 11.84LLKK120 pKa = 10.33EE121 pKa = 4.57KK122 pKa = 10.64IPDD125 pKa = 3.54KK126 pKa = 11.39VNLEE130 pKa = 5.01GIDD133 pKa = 3.5QHH135 pKa = 5.99HH136 pKa = 6.84TSAEE140 pKa = 4.23FIDD143 pKa = 4.69KK144 pKa = 10.75VGAPGGANQRR154 pKa = 11.84HH155 pKa = 5.3TPDD158 pKa = 2.67WMLPLILGLYY168 pKa = 10.42GDD170 pKa = 5.39LTPTWKK176 pKa = 10.43KK177 pKa = 9.4VVEE180 pKa = 4.3EE181 pKa = 4.52EE182 pKa = 4.17EE183 pKa = 5.07HH184 pKa = 5.63IHH186 pKa = 7.13GPSTKK191 pKa = 9.85KK192 pKa = 10.3KK193 pKa = 9.09PKK195 pKa = 8.72TSSPSSSTKK204 pKa = 9.24AAYY207 pKa = 9.21KK208 pKa = 9.81RR209 pKa = 11.84RR210 pKa = 11.84HH211 pKa = 5.67RR212 pKa = 11.84SARR215 pKa = 11.84PQNRR219 pKa = 11.84ARR221 pKa = 3.86
MM1 pKa = 7.49ALVPYY6 pKa = 9.8MPDD9 pKa = 3.09PDD11 pKa = 3.46IFFPGVSTFARR22 pKa = 11.84YY23 pKa = 9.82ANYY26 pKa = 10.46LDD28 pKa = 4.28PFHH31 pKa = 7.1WAPDD35 pKa = 3.59LYY37 pKa = 10.54HH38 pKa = 6.53QVSRR42 pKa = 11.84WIWEE46 pKa = 4.19SLMHH50 pKa = 6.47EE51 pKa = 4.53GRR53 pKa = 11.84RR54 pKa = 11.84QIGYY58 pKa = 10.03AGQEE62 pKa = 4.04LVRR65 pKa = 11.84YY66 pKa = 7.48TSTTVQDD73 pKa = 3.22TLARR77 pKa = 11.84FFEE80 pKa = 4.17NARR83 pKa = 11.84WAVSHH88 pKa = 6.96FSTNIYY94 pKa = 9.89TSLHH98 pKa = 6.17DD99 pKa = 4.42YY100 pKa = 10.5YY101 pKa = 11.3RR102 pKa = 11.84EE103 pKa = 3.99LPKK106 pKa = 10.7LKK108 pKa = 10.23PEE110 pKa = 4.03QARR113 pKa = 11.84QLSRR117 pKa = 11.84LLKK120 pKa = 10.33EE121 pKa = 4.57KK122 pKa = 10.64IPDD125 pKa = 3.54KK126 pKa = 11.39VNLEE130 pKa = 5.01GIDD133 pKa = 3.5QHH135 pKa = 5.99HH136 pKa = 6.84TSAEE140 pKa = 4.23FIDD143 pKa = 4.69KK144 pKa = 10.75VGAPGGANQRR154 pKa = 11.84HH155 pKa = 5.3TPDD158 pKa = 2.67WMLPLILGLYY168 pKa = 10.42GDD170 pKa = 5.39LTPTWKK176 pKa = 10.43KK177 pKa = 9.4VVEE180 pKa = 4.3EE181 pKa = 4.52EE182 pKa = 4.17EE183 pKa = 5.07HH184 pKa = 5.63IHH186 pKa = 7.13GPSTKK191 pKa = 9.85KK192 pKa = 10.3KK193 pKa = 9.09PKK195 pKa = 8.72TSSPSSSTKK204 pKa = 9.24AAYY207 pKa = 9.21KK208 pKa = 9.81RR209 pKa = 11.84RR210 pKa = 11.84HH211 pKa = 5.67RR212 pKa = 11.84SARR215 pKa = 11.84PQNRR219 pKa = 11.84ARR221 pKa = 3.86
Molecular weight: 25.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1781 |
175 |
684 |
356.2 |
40.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.019 ± 1.241 | 2.302 ± 0.861 |
5.222 ± 0.415 | 6.794 ± 0.518 |
3.762 ± 0.317 | 6.008 ± 0.844 |
2.864 ± 0.466 | 3.93 ± 0.434 |
6.569 ± 0.619 | 9.994 ± 0.583 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.92 ± 0.456 | 4.099 ± 0.592 |
6.008 ± 0.781 | 4.38 ± 0.335 |
4.773 ± 0.576 | 6.176 ± 0.524 |
5.671 ± 0.446 | 6.064 ± 0.646 |
1.853 ± 0.299 | 3.593 ± 0.38 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
