
Bat Paramyxovirus Eid_hel/GH-M74a/GHA/2009
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Henipavirus; Ghanaian bat henipavirus
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
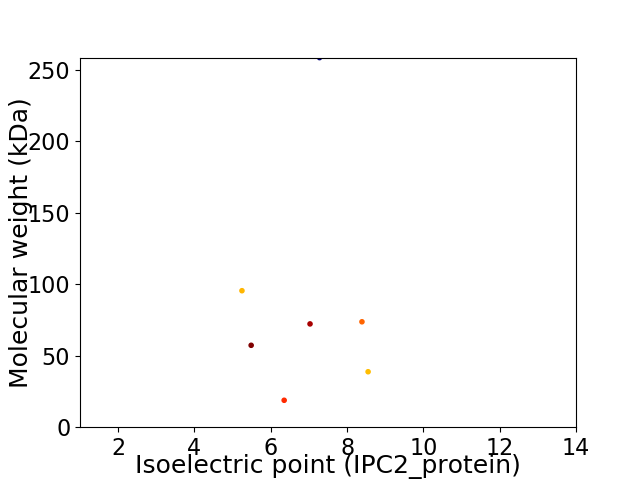
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I0E090|I0E090_9MONO Uncharacterized protein OS=Bat Paramyxovirus Eid_hel/GH-M74a/GHA/2009 OX=1112631 GN=C PE=4 SV=1
MM1 pKa = 7.94DD2 pKa = 5.25KK3 pKa = 10.97EE4 pKa = 4.41SLVDD8 pKa = 3.57NGIKK12 pKa = 9.82IIEE15 pKa = 4.93FIQKK19 pKa = 10.15NKK21 pKa = 11.08DD22 pKa = 3.7EE23 pKa = 4.19IQKK26 pKa = 9.59TYY28 pKa = 10.63GRR30 pKa = 11.84SQIQEE35 pKa = 3.5PRR37 pKa = 11.84TRR39 pKa = 11.84EE40 pKa = 3.36RR41 pKa = 11.84AAAWEE46 pKa = 3.91LFIRR50 pKa = 11.84GEE52 pKa = 4.11NPRR55 pKa = 11.84PEE57 pKa = 4.81GDD59 pKa = 2.96RR60 pKa = 11.84MAGVDD65 pKa = 3.44QQDD68 pKa = 3.46PTEE71 pKa = 4.41RR72 pKa = 11.84RR73 pKa = 11.84DD74 pKa = 3.57DD75 pKa = 4.01DD76 pKa = 3.98GRR78 pKa = 11.84PNTLARR84 pKa = 11.84DD85 pKa = 4.04GEE87 pKa = 4.38IGEE90 pKa = 4.68SGPNYY95 pKa = 9.84ISWSSGGDD103 pKa = 3.84DD104 pKa = 3.33IQLGPMVQDD113 pKa = 5.5FEE115 pKa = 6.01IDD117 pKa = 3.51DD118 pKa = 4.18NGIKK122 pKa = 10.43LPNSRR127 pKa = 11.84GSKK130 pKa = 9.04SAPRR134 pKa = 11.84CRR136 pKa = 11.84SVDD139 pKa = 3.15KK140 pKa = 11.16SRR142 pKa = 11.84ASDD145 pKa = 3.53WEE147 pKa = 3.91RR148 pKa = 11.84SPASNIDD155 pKa = 3.38HH156 pKa = 7.3LSNLSGGGKK165 pKa = 10.04DD166 pKa = 3.06LGNYY170 pKa = 8.22IKK172 pKa = 10.81NVGVMVYY179 pKa = 9.25PPTGAGVDD187 pKa = 3.7RR188 pKa = 11.84NSLTKK193 pKa = 9.84PVSGVRR199 pKa = 11.84QSRR202 pKa = 11.84LSKK205 pKa = 9.65LTWTKK210 pKa = 10.65EE211 pKa = 4.09SPLDD215 pKa = 3.59QPEE218 pKa = 4.5TVPEE222 pKa = 4.25TLNDD226 pKa = 3.41AYY228 pKa = 10.85VIKK231 pKa = 10.31EE232 pKa = 3.92ATSKK236 pKa = 11.27KK237 pKa = 10.17EE238 pKa = 3.72VTPEE242 pKa = 3.84DD243 pKa = 4.74GISILNPNAASFTPRR258 pKa = 11.84STPAPKK264 pKa = 10.19SVEE267 pKa = 3.87KK268 pKa = 10.6EE269 pKa = 3.81PNQRR273 pKa = 11.84QPAGGHH279 pKa = 6.52PEE281 pKa = 4.59DD282 pKa = 4.45KK283 pKa = 10.1GTPNRR288 pKa = 11.84SEE290 pKa = 4.04NPTTKK295 pKa = 9.45STAIKK300 pKa = 9.63NSKK303 pKa = 10.27VITKK307 pKa = 10.63AMVHH311 pKa = 5.97TSPNEE316 pKa = 3.68INQSARR322 pKa = 11.84SMNIEE327 pKa = 4.63FEE329 pKa = 4.49PKK331 pKa = 10.25SNPSTLTLEE340 pKa = 4.45TVQPSNQQTNNTPASHH356 pKa = 7.41PSGKK360 pKa = 6.65TTRR363 pKa = 11.84EE364 pKa = 4.08LPKK367 pKa = 10.47GAQPQSGTSRR377 pKa = 11.84AANIKK382 pKa = 10.76DD383 pKa = 3.41PTTAPNKK390 pKa = 9.8TSIQTKK396 pKa = 7.43PARR399 pKa = 11.84NASGGDD405 pKa = 3.34GTNPQMASPPPPPVVTNPPLKK426 pKa = 10.48KK427 pKa = 9.83RR428 pKa = 11.84VPKK431 pKa = 10.57LSEE434 pKa = 4.21PNTRR438 pKa = 11.84DD439 pKa = 3.65HH440 pKa = 7.1LSHH443 pKa = 6.69PTDD446 pKa = 3.26EE447 pKa = 4.82KK448 pKa = 10.8RR449 pKa = 11.84DD450 pKa = 3.66HH451 pKa = 6.87GGPAVKK457 pKa = 10.3SKK459 pKa = 8.58TQAHH463 pKa = 6.72RR464 pKa = 11.84GLNADD469 pKa = 3.51NTQNCTDD476 pKa = 3.41QPTPSSNKK484 pKa = 8.99AQPPKK489 pKa = 10.05TPKK492 pKa = 10.12PEE494 pKa = 4.0NPQAEE499 pKa = 4.75PKK501 pKa = 9.2PKK503 pKa = 10.2KK504 pKa = 9.77HH505 pKa = 6.5VSFPNLDD512 pKa = 3.43LTIEE516 pKa = 4.34EE517 pKa = 4.43EE518 pKa = 4.21GGTAVDD524 pKa = 3.14IMVNEE529 pKa = 4.17TRR531 pKa = 11.84EE532 pKa = 4.12GVIDD536 pKa = 3.27VHH538 pKa = 7.41LIKK541 pKa = 10.58EE542 pKa = 4.22DD543 pKa = 3.71NKK545 pKa = 10.34ILNMDD550 pKa = 3.6QDD552 pKa = 4.58NPCSNQITSQQEE564 pKa = 3.81TGVTKK569 pKa = 10.61RR570 pKa = 11.84GIGEE574 pKa = 3.94NSSCIGTKK582 pKa = 9.73EE583 pKa = 4.76DD584 pKa = 3.45SRR586 pKa = 11.84QLSGAIQSAQKK597 pKa = 10.52SRR599 pKa = 11.84LSQRR603 pKa = 11.84QEE605 pKa = 3.6NASAEE610 pKa = 4.31SARR613 pKa = 11.84PGVTSVPSDD622 pKa = 3.48DD623 pKa = 4.41PEE625 pKa = 4.8SEE627 pKa = 4.06EE628 pKa = 4.05VKK630 pKa = 10.05TYY632 pKa = 10.26RR633 pKa = 11.84ASDD636 pKa = 4.16AIIEE640 pKa = 4.01QLNNDD645 pKa = 3.92EE646 pKa = 4.4AQDD649 pKa = 3.96YY650 pKa = 11.3YY651 pKa = 11.95SFLDD655 pKa = 3.79LMEE658 pKa = 4.64NTDD661 pKa = 5.61DD662 pKa = 5.85DD663 pKa = 5.44DD664 pKa = 5.81LICEE668 pKa = 4.11EE669 pKa = 4.82TKK671 pKa = 10.71FSLLNTARR679 pKa = 11.84LISLNSRR686 pKa = 11.84IEE688 pKa = 4.24KK689 pKa = 10.06IEE691 pKa = 3.92EE692 pKa = 3.99QIKK695 pKa = 10.21KK696 pKa = 9.49IPAMEE701 pKa = 4.6KK702 pKa = 10.18KK703 pKa = 10.69LSDD706 pKa = 3.69IEE708 pKa = 4.32KK709 pKa = 10.79LLLKK713 pKa = 10.06TNTALSTIEE722 pKa = 3.82GHH724 pKa = 5.73ITSMMIMVPGKK735 pKa = 8.11TVNEE739 pKa = 4.11GEE741 pKa = 4.16IQINEE746 pKa = 3.91QLKK749 pKa = 9.89PVIGRR754 pKa = 11.84KK755 pKa = 9.93DD756 pKa = 3.35GVYY759 pKa = 10.98DD760 pKa = 3.97KK761 pKa = 10.6MILPRR766 pKa = 11.84VDD768 pKa = 3.45KK769 pKa = 10.75KK770 pKa = 10.97ISLVTDD776 pKa = 4.14RR777 pKa = 11.84TPLCDD782 pKa = 4.89DD783 pKa = 4.45FKK785 pKa = 11.22EE786 pKa = 4.39DD787 pKa = 4.46PYY789 pKa = 11.85VLNEE793 pKa = 4.28KK794 pKa = 10.59LILDD798 pKa = 5.27PINPSVTNASAFIPSYY814 pKa = 11.43DD815 pKa = 3.9NISPAVLRR823 pKa = 11.84SLIRR827 pKa = 11.84SNVEE831 pKa = 3.52DD832 pKa = 3.74RR833 pKa = 11.84EE834 pKa = 4.44TRR836 pKa = 11.84VEE838 pKa = 4.69LIEE841 pKa = 5.06LVNQARR847 pKa = 11.84NDD849 pKa = 3.71SEE851 pKa = 4.76LNEE854 pKa = 3.95ILALVNDD861 pKa = 5.38IIDD864 pKa = 4.05SNQSGVV870 pKa = 3.08
MM1 pKa = 7.94DD2 pKa = 5.25KK3 pKa = 10.97EE4 pKa = 4.41SLVDD8 pKa = 3.57NGIKK12 pKa = 9.82IIEE15 pKa = 4.93FIQKK19 pKa = 10.15NKK21 pKa = 11.08DD22 pKa = 3.7EE23 pKa = 4.19IQKK26 pKa = 9.59TYY28 pKa = 10.63GRR30 pKa = 11.84SQIQEE35 pKa = 3.5PRR37 pKa = 11.84TRR39 pKa = 11.84EE40 pKa = 3.36RR41 pKa = 11.84AAAWEE46 pKa = 3.91LFIRR50 pKa = 11.84GEE52 pKa = 4.11NPRR55 pKa = 11.84PEE57 pKa = 4.81GDD59 pKa = 2.96RR60 pKa = 11.84MAGVDD65 pKa = 3.44QQDD68 pKa = 3.46PTEE71 pKa = 4.41RR72 pKa = 11.84RR73 pKa = 11.84DD74 pKa = 3.57DD75 pKa = 4.01DD76 pKa = 3.98GRR78 pKa = 11.84PNTLARR84 pKa = 11.84DD85 pKa = 4.04GEE87 pKa = 4.38IGEE90 pKa = 4.68SGPNYY95 pKa = 9.84ISWSSGGDD103 pKa = 3.84DD104 pKa = 3.33IQLGPMVQDD113 pKa = 5.5FEE115 pKa = 6.01IDD117 pKa = 3.51DD118 pKa = 4.18NGIKK122 pKa = 10.43LPNSRR127 pKa = 11.84GSKK130 pKa = 9.04SAPRR134 pKa = 11.84CRR136 pKa = 11.84SVDD139 pKa = 3.15KK140 pKa = 11.16SRR142 pKa = 11.84ASDD145 pKa = 3.53WEE147 pKa = 3.91RR148 pKa = 11.84SPASNIDD155 pKa = 3.38HH156 pKa = 7.3LSNLSGGGKK165 pKa = 10.04DD166 pKa = 3.06LGNYY170 pKa = 8.22IKK172 pKa = 10.81NVGVMVYY179 pKa = 9.25PPTGAGVDD187 pKa = 3.7RR188 pKa = 11.84NSLTKK193 pKa = 9.84PVSGVRR199 pKa = 11.84QSRR202 pKa = 11.84LSKK205 pKa = 9.65LTWTKK210 pKa = 10.65EE211 pKa = 4.09SPLDD215 pKa = 3.59QPEE218 pKa = 4.5TVPEE222 pKa = 4.25TLNDD226 pKa = 3.41AYY228 pKa = 10.85VIKK231 pKa = 10.31EE232 pKa = 3.92ATSKK236 pKa = 11.27KK237 pKa = 10.17EE238 pKa = 3.72VTPEE242 pKa = 3.84DD243 pKa = 4.74GISILNPNAASFTPRR258 pKa = 11.84STPAPKK264 pKa = 10.19SVEE267 pKa = 3.87KK268 pKa = 10.6EE269 pKa = 3.81PNQRR273 pKa = 11.84QPAGGHH279 pKa = 6.52PEE281 pKa = 4.59DD282 pKa = 4.45KK283 pKa = 10.1GTPNRR288 pKa = 11.84SEE290 pKa = 4.04NPTTKK295 pKa = 9.45STAIKK300 pKa = 9.63NSKK303 pKa = 10.27VITKK307 pKa = 10.63AMVHH311 pKa = 5.97TSPNEE316 pKa = 3.68INQSARR322 pKa = 11.84SMNIEE327 pKa = 4.63FEE329 pKa = 4.49PKK331 pKa = 10.25SNPSTLTLEE340 pKa = 4.45TVQPSNQQTNNTPASHH356 pKa = 7.41PSGKK360 pKa = 6.65TTRR363 pKa = 11.84EE364 pKa = 4.08LPKK367 pKa = 10.47GAQPQSGTSRR377 pKa = 11.84AANIKK382 pKa = 10.76DD383 pKa = 3.41PTTAPNKK390 pKa = 9.8TSIQTKK396 pKa = 7.43PARR399 pKa = 11.84NASGGDD405 pKa = 3.34GTNPQMASPPPPPVVTNPPLKK426 pKa = 10.48KK427 pKa = 9.83RR428 pKa = 11.84VPKK431 pKa = 10.57LSEE434 pKa = 4.21PNTRR438 pKa = 11.84DD439 pKa = 3.65HH440 pKa = 7.1LSHH443 pKa = 6.69PTDD446 pKa = 3.26EE447 pKa = 4.82KK448 pKa = 10.8RR449 pKa = 11.84DD450 pKa = 3.66HH451 pKa = 6.87GGPAVKK457 pKa = 10.3SKK459 pKa = 8.58TQAHH463 pKa = 6.72RR464 pKa = 11.84GLNADD469 pKa = 3.51NTQNCTDD476 pKa = 3.41QPTPSSNKK484 pKa = 8.99AQPPKK489 pKa = 10.05TPKK492 pKa = 10.12PEE494 pKa = 4.0NPQAEE499 pKa = 4.75PKK501 pKa = 9.2PKK503 pKa = 10.2KK504 pKa = 9.77HH505 pKa = 6.5VSFPNLDD512 pKa = 3.43LTIEE516 pKa = 4.34EE517 pKa = 4.43EE518 pKa = 4.21GGTAVDD524 pKa = 3.14IMVNEE529 pKa = 4.17TRR531 pKa = 11.84EE532 pKa = 4.12GVIDD536 pKa = 3.27VHH538 pKa = 7.41LIKK541 pKa = 10.58EE542 pKa = 4.22DD543 pKa = 3.71NKK545 pKa = 10.34ILNMDD550 pKa = 3.6QDD552 pKa = 4.58NPCSNQITSQQEE564 pKa = 3.81TGVTKK569 pKa = 10.61RR570 pKa = 11.84GIGEE574 pKa = 3.94NSSCIGTKK582 pKa = 9.73EE583 pKa = 4.76DD584 pKa = 3.45SRR586 pKa = 11.84QLSGAIQSAQKK597 pKa = 10.52SRR599 pKa = 11.84LSQRR603 pKa = 11.84QEE605 pKa = 3.6NASAEE610 pKa = 4.31SARR613 pKa = 11.84PGVTSVPSDD622 pKa = 3.48DD623 pKa = 4.41PEE625 pKa = 4.8SEE627 pKa = 4.06EE628 pKa = 4.05VKK630 pKa = 10.05TYY632 pKa = 10.26RR633 pKa = 11.84ASDD636 pKa = 4.16AIIEE640 pKa = 4.01QLNNDD645 pKa = 3.92EE646 pKa = 4.4AQDD649 pKa = 3.96YY650 pKa = 11.3YY651 pKa = 11.95SFLDD655 pKa = 3.79LMEE658 pKa = 4.64NTDD661 pKa = 5.61DD662 pKa = 5.85DD663 pKa = 5.44DD664 pKa = 5.81LICEE668 pKa = 4.11EE669 pKa = 4.82TKK671 pKa = 10.71FSLLNTARR679 pKa = 11.84LISLNSRR686 pKa = 11.84IEE688 pKa = 4.24KK689 pKa = 10.06IEE691 pKa = 3.92EE692 pKa = 3.99QIKK695 pKa = 10.21KK696 pKa = 9.49IPAMEE701 pKa = 4.6KK702 pKa = 10.18KK703 pKa = 10.69LSDD706 pKa = 3.69IEE708 pKa = 4.32KK709 pKa = 10.79LLLKK713 pKa = 10.06TNTALSTIEE722 pKa = 3.82GHH724 pKa = 5.73ITSMMIMVPGKK735 pKa = 8.11TVNEE739 pKa = 4.11GEE741 pKa = 4.16IQINEE746 pKa = 3.91QLKK749 pKa = 9.89PVIGRR754 pKa = 11.84KK755 pKa = 9.93DD756 pKa = 3.35GVYY759 pKa = 10.98DD760 pKa = 3.97KK761 pKa = 10.6MILPRR766 pKa = 11.84VDD768 pKa = 3.45KK769 pKa = 10.75KK770 pKa = 10.97ISLVTDD776 pKa = 4.14RR777 pKa = 11.84TPLCDD782 pKa = 4.89DD783 pKa = 4.45FKK785 pKa = 11.22EE786 pKa = 4.39DD787 pKa = 4.46PYY789 pKa = 11.85VLNEE793 pKa = 4.28KK794 pKa = 10.59LILDD798 pKa = 5.27PINPSVTNASAFIPSYY814 pKa = 11.43DD815 pKa = 3.9NISPAVLRR823 pKa = 11.84SLIRR827 pKa = 11.84SNVEE831 pKa = 3.52DD832 pKa = 3.74RR833 pKa = 11.84EE834 pKa = 4.44TRR836 pKa = 11.84VEE838 pKa = 4.69LIEE841 pKa = 5.06LVNQARR847 pKa = 11.84NDD849 pKa = 3.71SEE851 pKa = 4.76LNEE854 pKa = 3.95ILALVNDD861 pKa = 5.38IIDD864 pKa = 4.05SNQSGVV870 pKa = 3.08
Molecular weight: 95.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I0E092|I0E092_9MONO Fusion glycoprotein F0 OS=Bat Paramyxovirus Eid_hel/GH-M74a/GHA/2009 OX=1112631 GN=F PE=3 SV=1
MM1 pKa = 7.25EE2 pKa = 6.66AITNFMPYY10 pKa = 7.5TWEE13 pKa = 4.35FGGSLNKK20 pKa = 9.69VEE22 pKa = 5.3PEE24 pKa = 3.66LDD26 pKa = 3.78EE27 pKa = 4.7NGSMIPKK34 pKa = 9.6VRR36 pKa = 11.84VLNPGYY42 pKa = 10.43NDD44 pKa = 3.74RR45 pKa = 11.84KK46 pKa = 10.09SSGYY50 pKa = 10.21QYY52 pKa = 10.16LICYY56 pKa = 8.97GFVEE60 pKa = 5.43DD61 pKa = 5.92IRR63 pKa = 11.84SEE65 pKa = 4.41SNGDD69 pKa = 3.24KK70 pKa = 10.71KK71 pKa = 10.37PTIRR75 pKa = 11.84TTASFPLGVGRR86 pKa = 11.84SDD88 pKa = 3.9ASPEE92 pKa = 3.83NLLDD96 pKa = 4.02EE97 pKa = 4.45VCKK100 pKa = 10.91LSITVRR106 pKa = 11.84RR107 pKa = 11.84TSGAEE112 pKa = 3.56EE113 pKa = 4.26KK114 pKa = 10.25IVYY117 pKa = 9.16GVSGGILYY125 pKa = 10.24LDD127 pKa = 3.59PWKK130 pKa = 10.75KK131 pKa = 9.98VLKK134 pKa = 10.51NGAIFTATKK143 pKa = 8.33VCRR146 pKa = 11.84NIEE149 pKa = 4.11CVLVDD154 pKa = 2.93KK155 pKa = 10.7HH156 pKa = 5.78QSLRR160 pKa = 11.84IFFLSITKK168 pKa = 8.83LTDD171 pKa = 2.71RR172 pKa = 11.84GYY174 pKa = 8.51YY175 pKa = 8.3TIPRR179 pKa = 11.84RR180 pKa = 11.84MLEE183 pKa = 3.61FRR185 pKa = 11.84YY186 pKa = 10.9NNAIALNLLVTLAISTDD203 pKa = 3.48TTKK206 pKa = 11.0SGVRR210 pKa = 11.84GIKK213 pKa = 10.43NEE215 pKa = 4.22DD216 pKa = 3.4GFTLVTFMTHH226 pKa = 5.24MGNFQRR232 pKa = 11.84RR233 pKa = 11.84AGKK236 pKa = 8.44TYY238 pKa = 10.29SLEE241 pKa = 4.02YY242 pKa = 10.3CSRR245 pKa = 11.84KK246 pKa = 8.19VDD248 pKa = 3.32KK249 pKa = 10.64MKK251 pKa = 10.56MKK253 pKa = 10.57FSLGAIGGLSFHH265 pKa = 6.03VRR267 pKa = 11.84IDD269 pKa = 3.57GVISKK274 pKa = 10.54RR275 pKa = 11.84LFAEE279 pKa = 3.66MGFRR283 pKa = 11.84RR284 pKa = 11.84NICYY288 pKa = 10.22CLIDD292 pKa = 3.7INPWINKK299 pKa = 6.94LTWSNTCEE307 pKa = 3.95IRR309 pKa = 11.84NVAAVLQPSVPRR321 pKa = 11.84DD322 pKa = 3.04FMIYY326 pKa = 10.67DD327 pKa = 4.21DD328 pKa = 4.75VFIDD332 pKa = 3.62NTGKK336 pKa = 10.27ILPSKK341 pKa = 10.14HH342 pKa = 5.56AA343 pKa = 4.26
MM1 pKa = 7.25EE2 pKa = 6.66AITNFMPYY10 pKa = 7.5TWEE13 pKa = 4.35FGGSLNKK20 pKa = 9.69VEE22 pKa = 5.3PEE24 pKa = 3.66LDD26 pKa = 3.78EE27 pKa = 4.7NGSMIPKK34 pKa = 9.6VRR36 pKa = 11.84VLNPGYY42 pKa = 10.43NDD44 pKa = 3.74RR45 pKa = 11.84KK46 pKa = 10.09SSGYY50 pKa = 10.21QYY52 pKa = 10.16LICYY56 pKa = 8.97GFVEE60 pKa = 5.43DD61 pKa = 5.92IRR63 pKa = 11.84SEE65 pKa = 4.41SNGDD69 pKa = 3.24KK70 pKa = 10.71KK71 pKa = 10.37PTIRR75 pKa = 11.84TTASFPLGVGRR86 pKa = 11.84SDD88 pKa = 3.9ASPEE92 pKa = 3.83NLLDD96 pKa = 4.02EE97 pKa = 4.45VCKK100 pKa = 10.91LSITVRR106 pKa = 11.84RR107 pKa = 11.84TSGAEE112 pKa = 3.56EE113 pKa = 4.26KK114 pKa = 10.25IVYY117 pKa = 9.16GVSGGILYY125 pKa = 10.24LDD127 pKa = 3.59PWKK130 pKa = 10.75KK131 pKa = 9.98VLKK134 pKa = 10.51NGAIFTATKK143 pKa = 8.33VCRR146 pKa = 11.84NIEE149 pKa = 4.11CVLVDD154 pKa = 2.93KK155 pKa = 10.7HH156 pKa = 5.78QSLRR160 pKa = 11.84IFFLSITKK168 pKa = 8.83LTDD171 pKa = 2.71RR172 pKa = 11.84GYY174 pKa = 8.51YY175 pKa = 8.3TIPRR179 pKa = 11.84RR180 pKa = 11.84MLEE183 pKa = 3.61FRR185 pKa = 11.84YY186 pKa = 10.9NNAIALNLLVTLAISTDD203 pKa = 3.48TTKK206 pKa = 11.0SGVRR210 pKa = 11.84GIKK213 pKa = 10.43NEE215 pKa = 4.22DD216 pKa = 3.4GFTLVTFMTHH226 pKa = 5.24MGNFQRR232 pKa = 11.84RR233 pKa = 11.84AGKK236 pKa = 8.44TYY238 pKa = 10.29SLEE241 pKa = 4.02YY242 pKa = 10.3CSRR245 pKa = 11.84KK246 pKa = 8.19VDD248 pKa = 3.32KK249 pKa = 10.64MKK251 pKa = 10.56MKK253 pKa = 10.57FSLGAIGGLSFHH265 pKa = 6.03VRR267 pKa = 11.84IDD269 pKa = 3.57GVISKK274 pKa = 10.54RR275 pKa = 11.84LFAEE279 pKa = 3.66MGFRR283 pKa = 11.84RR284 pKa = 11.84NICYY288 pKa = 10.22CLIDD292 pKa = 3.7INPWINKK299 pKa = 6.94LTWSNTCEE307 pKa = 3.95IRR309 pKa = 11.84NVAAVLQPSVPRR321 pKa = 11.84DD322 pKa = 3.04FMIYY326 pKa = 10.67DD327 pKa = 4.21DD328 pKa = 4.75VFIDD332 pKa = 3.62NTGKK336 pKa = 10.27ILPSKK341 pKa = 10.14HH342 pKa = 5.56AA343 pKa = 4.26
Molecular weight: 38.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5433 |
162 |
2250 |
776.1 |
87.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.822 ± 0.521 | 1.546 ± 0.305 |
5.393 ± 0.315 | 6.203 ± 0.442 |
3.258 ± 0.586 | 5.154 ± 0.446 |
1.859 ± 0.358 | 8.448 ± 0.674 |
6.35 ± 0.438 | 9.074 ± 0.648 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.558 ± 0.217 | 6.037 ± 0.438 |
4.859 ± 0.762 | 3.792 ± 0.298 |
5.503 ± 0.4 | 8.32 ± 0.308 |
6.534 ± 0.563 | 5.54 ± 0.142 |
0.92 ± 0.152 | 3.828 ± 0.583 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
