
Leptolyngbya boryana NIES-2135
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Leptolyngbyaceae; Leptolyngbya; Leptolyngbya boryana
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
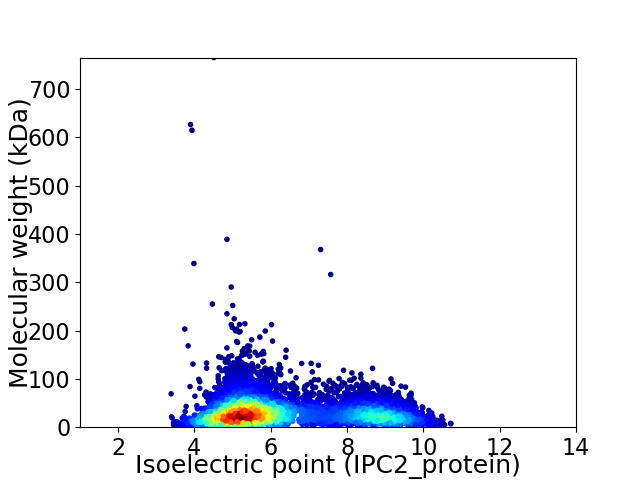
Virtual 2D-PAGE plot for 6678 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z4JQA1|A0A1Z4JQA1_LEPBY Cell wall hydrolase/autolysin OS=Leptolyngbya boryana NIES-2135 OX=1973484 GN=NIES2135_57640 PE=4 SV=1
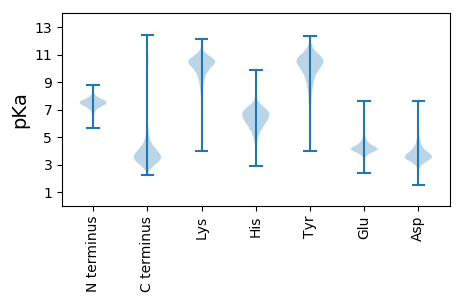
MM1 pKa = 7.09ATVGLFYY8 pKa = 10.15GTQTGNTEE16 pKa = 4.45TIAEE20 pKa = 5.17AIQAQLGGDD29 pKa = 4.15SIVEE33 pKa = 3.94LSDD36 pKa = 2.99IANASSEE43 pKa = 4.11DD44 pKa = 3.79FEE46 pKa = 4.87PYY48 pKa = 9.72EE49 pKa = 4.15YY50 pKa = 10.78LIIGCPTWNIGEE62 pKa = 4.5LQADD66 pKa = 3.44WEE68 pKa = 4.51GFYY71 pKa = 11.3DD72 pKa = 4.33EE73 pKa = 5.75LDD75 pKa = 3.83NINFNGKK82 pKa = 8.53KK83 pKa = 9.33VAYY86 pKa = 8.86FGCGDD91 pKa = 3.67QIGYY95 pKa = 10.75ADD97 pKa = 4.11NFQDD101 pKa = 4.12AMGILEE107 pKa = 4.46EE108 pKa = 5.13KK109 pKa = 10.48ISGLGGQTVGMTSTAGYY126 pKa = 8.88EE127 pKa = 4.21HH128 pKa = 6.69QEE130 pKa = 4.17SKK132 pKa = 10.97AVRR135 pKa = 11.84GDD137 pKa = 3.64KK138 pKa = 10.74FCGLAIDD145 pKa = 5.14EE146 pKa = 5.32DD147 pKa = 4.18NQSDD151 pKa = 4.0LTDD154 pKa = 3.49EE155 pKa = 5.26RR156 pKa = 11.84IKK158 pKa = 10.99AWATQLKK165 pKa = 9.89SAFGII170 pKa = 4.18
MM1 pKa = 7.09ATVGLFYY8 pKa = 10.15GTQTGNTEE16 pKa = 4.45TIAEE20 pKa = 5.17AIQAQLGGDD29 pKa = 4.15SIVEE33 pKa = 3.94LSDD36 pKa = 2.99IANASSEE43 pKa = 4.11DD44 pKa = 3.79FEE46 pKa = 4.87PYY48 pKa = 9.72EE49 pKa = 4.15YY50 pKa = 10.78LIIGCPTWNIGEE62 pKa = 4.5LQADD66 pKa = 3.44WEE68 pKa = 4.51GFYY71 pKa = 11.3DD72 pKa = 4.33EE73 pKa = 5.75LDD75 pKa = 3.83NINFNGKK82 pKa = 8.53KK83 pKa = 9.33VAYY86 pKa = 8.86FGCGDD91 pKa = 3.67QIGYY95 pKa = 10.75ADD97 pKa = 4.11NFQDD101 pKa = 4.12AMGILEE107 pKa = 4.46EE108 pKa = 5.13KK109 pKa = 10.48ISGLGGQTVGMTSTAGYY126 pKa = 8.88EE127 pKa = 4.21HH128 pKa = 6.69QEE130 pKa = 4.17SKK132 pKa = 10.97AVRR135 pKa = 11.84GDD137 pKa = 3.64KK138 pKa = 10.74FCGLAIDD145 pKa = 5.14EE146 pKa = 5.32DD147 pKa = 4.18NQSDD151 pKa = 4.0LTDD154 pKa = 3.49EE155 pKa = 5.26RR156 pKa = 11.84IKK158 pKa = 10.99AWATQLKK165 pKa = 9.89SAFGII170 pKa = 4.18
Molecular weight: 18.44 kDa
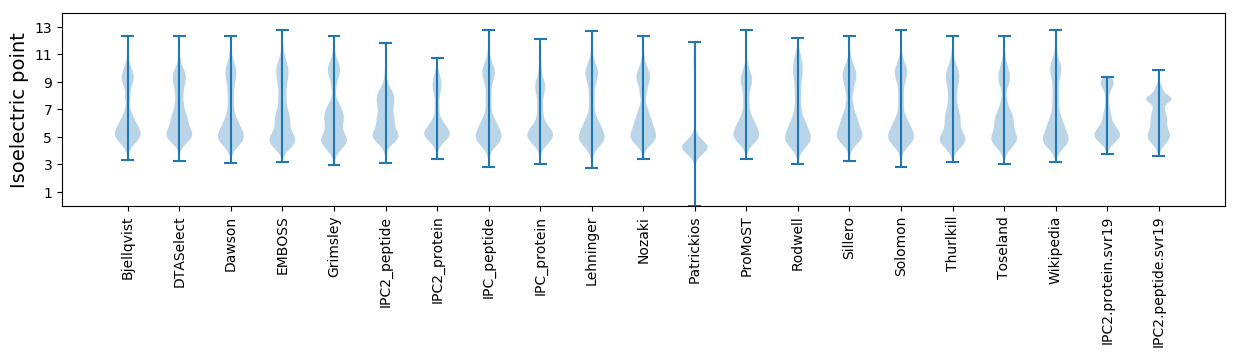
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z4JMH1|A0A1Z4JMH1_LEPBY Uncharacterized protein OS=Leptolyngbya boryana NIES-2135 OX=1973484 GN=NIES2135_47190 PE=4 SV=1
MM1 pKa = 8.04LDD3 pKa = 4.08LLLMASVGFLGSFGHH18 pKa = 6.47CLGMCSPLTVAFSLSNDD35 pKa = 3.19RR36 pKa = 11.84TSRR39 pKa = 11.84FAQLRR44 pKa = 11.84FNLLLNAGRR53 pKa = 11.84VMSYY57 pKa = 9.08TLVGAAIGALGSVLIAGGQLAGIDD81 pKa = 3.46SALRR85 pKa = 11.84RR86 pKa = 11.84GLAIVTGLLLIWFGLANINPTWLPQVPILHH116 pKa = 7.16PIQGKK121 pKa = 8.55LHH123 pKa = 6.84DD124 pKa = 4.86RR125 pKa = 11.84LSQRR129 pKa = 11.84LGDD132 pKa = 3.86FSRR135 pKa = 11.84SRR137 pKa = 11.84KK138 pKa = 6.53WWTPAILGLVWGLIPCGFLYY158 pKa = 10.61AAQIKK163 pKa = 9.76AAEE166 pKa = 4.4TGNALQGALTMLAFGGGTAPMMISTGLFAGALSRR200 pKa = 11.84DD201 pKa = 3.39RR202 pKa = 11.84RR203 pKa = 11.84SQLFKK208 pKa = 10.43LAGWITLTIGLLTLFRR224 pKa = 11.84SSDD227 pKa = 3.34MVDD230 pKa = 3.7YY231 pKa = 10.0TGHH234 pKa = 7.01AALICLMVALIARR247 pKa = 11.84PISRR251 pKa = 11.84IWAAPLRR258 pKa = 11.84YY259 pKa = 9.12RR260 pKa = 11.84RR261 pKa = 11.84ALGVGAFVLSCAHH274 pKa = 6.4MGHH277 pKa = 6.51FLDD280 pKa = 5.84HH281 pKa = 6.12TFNWNIYY288 pKa = 9.89AIAFLLPNQQIGMWAGIIALGLMIPLALTSFDD320 pKa = 3.56FAVAKK325 pKa = 10.73LGTAWRR331 pKa = 11.84KK332 pKa = 8.23IHH334 pKa = 6.35LVSIPVLILAVIHH347 pKa = 6.17TSLTGSNYY355 pKa = 10.31LGEE358 pKa = 4.38INSDD362 pKa = 4.16FIHH365 pKa = 7.02RR366 pKa = 11.84FRR368 pKa = 11.84VGILIFLVITVLLIRR383 pKa = 11.84LQIFWSLFSLEE394 pKa = 4.14KK395 pKa = 10.31FYY397 pKa = 11.13AASKK401 pKa = 10.24RR402 pKa = 3.7
MM1 pKa = 8.04LDD3 pKa = 4.08LLLMASVGFLGSFGHH18 pKa = 6.47CLGMCSPLTVAFSLSNDD35 pKa = 3.19RR36 pKa = 11.84TSRR39 pKa = 11.84FAQLRR44 pKa = 11.84FNLLLNAGRR53 pKa = 11.84VMSYY57 pKa = 9.08TLVGAAIGALGSVLIAGGQLAGIDD81 pKa = 3.46SALRR85 pKa = 11.84RR86 pKa = 11.84GLAIVTGLLLIWFGLANINPTWLPQVPILHH116 pKa = 7.16PIQGKK121 pKa = 8.55LHH123 pKa = 6.84DD124 pKa = 4.86RR125 pKa = 11.84LSQRR129 pKa = 11.84LGDD132 pKa = 3.86FSRR135 pKa = 11.84SRR137 pKa = 11.84KK138 pKa = 6.53WWTPAILGLVWGLIPCGFLYY158 pKa = 10.61AAQIKK163 pKa = 9.76AAEE166 pKa = 4.4TGNALQGALTMLAFGGGTAPMMISTGLFAGALSRR200 pKa = 11.84DD201 pKa = 3.39RR202 pKa = 11.84RR203 pKa = 11.84SQLFKK208 pKa = 10.43LAGWITLTIGLLTLFRR224 pKa = 11.84SSDD227 pKa = 3.34MVDD230 pKa = 3.7YY231 pKa = 10.0TGHH234 pKa = 7.01AALICLMVALIARR247 pKa = 11.84PISRR251 pKa = 11.84IWAAPLRR258 pKa = 11.84YY259 pKa = 9.12RR260 pKa = 11.84RR261 pKa = 11.84ALGVGAFVLSCAHH274 pKa = 6.4MGHH277 pKa = 6.51FLDD280 pKa = 5.84HH281 pKa = 6.12TFNWNIYY288 pKa = 9.89AIAFLLPNQQIGMWAGIIALGLMIPLALTSFDD320 pKa = 3.56FAVAKK325 pKa = 10.73LGTAWRR331 pKa = 11.84KK332 pKa = 8.23IHH334 pKa = 6.35LVSIPVLILAVIHH347 pKa = 6.17TSLTGSNYY355 pKa = 10.31LGEE358 pKa = 4.38INSDD362 pKa = 4.16FIHH365 pKa = 7.02RR366 pKa = 11.84FRR368 pKa = 11.84VGILIFLVITVLLIRR383 pKa = 11.84LQIFWSLFSLEE394 pKa = 4.14KK395 pKa = 10.31FYY397 pKa = 11.13AASKK401 pKa = 10.24RR402 pKa = 3.7
Molecular weight: 43.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2078917 |
29 |
7046 |
311.3 |
34.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.975 ± 0.033 | 0.962 ± 0.012 |
5.072 ± 0.025 | 6.149 ± 0.036 |
3.944 ± 0.021 | 6.561 ± 0.039 |
1.901 ± 0.016 | 6.373 ± 0.022 |
4.034 ± 0.03 | 10.881 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.972 ± 0.015 | 3.688 ± 0.029 |
4.814 ± 0.03 | 5.651 ± 0.033 |
5.846 ± 0.028 | 6.602 ± 0.033 |
5.751 ± 0.04 | 6.619 ± 0.027 |
1.458 ± 0.014 | 2.744 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
