
Panicum streak virus (isolate Kenya) (PanSV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus; Panicum streak virus
Average proteome isoelectric point is 7.59
Get precalculated fractions of proteins
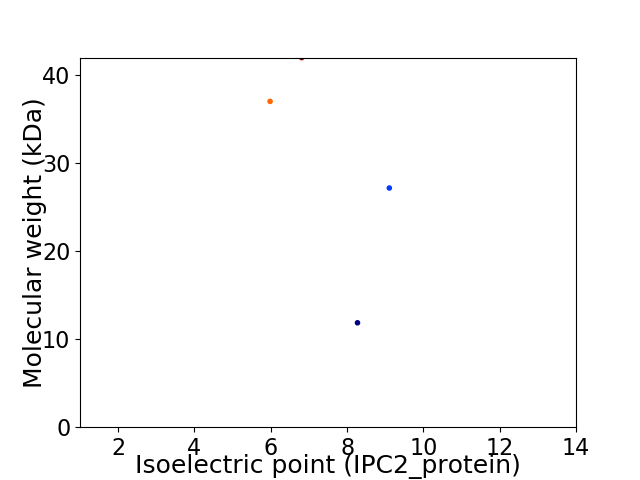
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q00338|REPA_PASVK Replication-associated protein A OS=Panicum streak virus (isolate Kenya) OX=268780 GN=C1 PE=3 SV=1
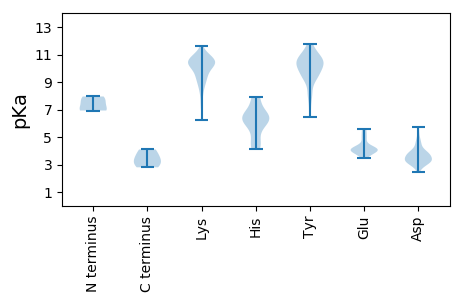
MM1 pKa = 6.93STVGSSSEE9 pKa = 3.68GRR11 pKa = 11.84HH12 pKa = 4.16SVRR15 pKa = 11.84CFRR18 pKa = 11.84HH19 pKa = 6.14RR20 pKa = 11.84NANTFLTYY28 pKa = 10.26SKK30 pKa = 10.88CPLEE34 pKa = 3.93PEE36 pKa = 4.86FIGEE40 pKa = 3.92HH41 pKa = 6.27LFRR44 pKa = 11.84LTRR47 pKa = 11.84EE48 pKa = 3.93YY49 pKa = 10.61EE50 pKa = 3.57PAYY53 pKa = 9.34ILVVRR58 pKa = 11.84EE59 pKa = 3.9THH61 pKa = 7.15TDD63 pKa = 3.88GTWHH67 pKa = 6.33CHH69 pKa = 6.64ALLQCIKK76 pKa = 10.22PCTTRR81 pKa = 11.84DD82 pKa = 3.17EE83 pKa = 4.79RR84 pKa = 11.84YY85 pKa = 9.6FDD87 pKa = 3.28IDD89 pKa = 3.79RR90 pKa = 11.84YY91 pKa = 10.29HH92 pKa = 7.89GNIQSAKK99 pKa = 8.26STDD102 pKa = 3.09KK103 pKa = 10.55VRR105 pKa = 11.84EE106 pKa = 4.1YY107 pKa = 10.68ILKK110 pKa = 10.31DD111 pKa = 3.71PKK113 pKa = 11.09DD114 pKa = 3.29KK115 pKa = 10.5WEE117 pKa = 4.22KK118 pKa = 8.82GTYY121 pKa = 9.25IPRR124 pKa = 11.84KK125 pKa = 9.51KK126 pKa = 10.78SFVPPGKK133 pKa = 10.29EE134 pKa = 3.49PAEE137 pKa = 4.14KK138 pKa = 10.46KK139 pKa = 7.32PTKK142 pKa = 10.27DD143 pKa = 2.81EE144 pKa = 3.99VMRR147 pKa = 11.84EE148 pKa = 3.94IMTHH152 pKa = 4.33ATSRR156 pKa = 11.84EE157 pKa = 4.06EE158 pKa = 3.93YY159 pKa = 10.39LSLVQSSLPYY169 pKa = 10.68DD170 pKa = 3.27WATKK174 pKa = 10.48LNYY177 pKa = 9.62FEE179 pKa = 5.23YY180 pKa = 10.2SASRR184 pKa = 11.84LFPDD188 pKa = 3.2IAEE191 pKa = 5.11PYY193 pKa = 7.99TNPHH197 pKa = 6.38PTTEE201 pKa = 4.23YY202 pKa = 10.94DD203 pKa = 3.4LHH205 pKa = 7.92CNEE208 pKa = 5.52TIEE211 pKa = 5.55DD212 pKa = 3.55WLKK215 pKa = 10.05PNIYY219 pKa = 9.58QVSPQAYY226 pKa = 8.42KK227 pKa = 10.88LLEE230 pKa = 4.21PSCLSLEE237 pKa = 4.11QAIADD242 pKa = 4.68LEE244 pKa = 4.34WLDD247 pKa = 3.51DD248 pKa = 3.85TTRR251 pKa = 11.84MLQEE255 pKa = 4.21KK256 pKa = 9.35EE257 pKa = 4.3RR258 pKa = 11.84EE259 pKa = 4.18ASTSAAQHH267 pKa = 5.69GQVKK271 pKa = 10.29HH272 pKa = 6.52PGLEE276 pKa = 4.15ASDD279 pKa = 3.77DD280 pKa = 3.82TTTGKK285 pKa = 9.01TISTGLHH292 pKa = 4.59MMKK295 pKa = 10.4KK296 pKa = 10.09LSTMSLTTFPSSSVRR311 pKa = 11.84AGNDD315 pKa = 2.47SSAAKK320 pKa = 8.85KK321 pKa = 6.24TTT323 pKa = 3.14
MM1 pKa = 6.93STVGSSSEE9 pKa = 3.68GRR11 pKa = 11.84HH12 pKa = 4.16SVRR15 pKa = 11.84CFRR18 pKa = 11.84HH19 pKa = 6.14RR20 pKa = 11.84NANTFLTYY28 pKa = 10.26SKK30 pKa = 10.88CPLEE34 pKa = 3.93PEE36 pKa = 4.86FIGEE40 pKa = 3.92HH41 pKa = 6.27LFRR44 pKa = 11.84LTRR47 pKa = 11.84EE48 pKa = 3.93YY49 pKa = 10.61EE50 pKa = 3.57PAYY53 pKa = 9.34ILVVRR58 pKa = 11.84EE59 pKa = 3.9THH61 pKa = 7.15TDD63 pKa = 3.88GTWHH67 pKa = 6.33CHH69 pKa = 6.64ALLQCIKK76 pKa = 10.22PCTTRR81 pKa = 11.84DD82 pKa = 3.17EE83 pKa = 4.79RR84 pKa = 11.84YY85 pKa = 9.6FDD87 pKa = 3.28IDD89 pKa = 3.79RR90 pKa = 11.84YY91 pKa = 10.29HH92 pKa = 7.89GNIQSAKK99 pKa = 8.26STDD102 pKa = 3.09KK103 pKa = 10.55VRR105 pKa = 11.84EE106 pKa = 4.1YY107 pKa = 10.68ILKK110 pKa = 10.31DD111 pKa = 3.71PKK113 pKa = 11.09DD114 pKa = 3.29KK115 pKa = 10.5WEE117 pKa = 4.22KK118 pKa = 8.82GTYY121 pKa = 9.25IPRR124 pKa = 11.84KK125 pKa = 9.51KK126 pKa = 10.78SFVPPGKK133 pKa = 10.29EE134 pKa = 3.49PAEE137 pKa = 4.14KK138 pKa = 10.46KK139 pKa = 7.32PTKK142 pKa = 10.27DD143 pKa = 2.81EE144 pKa = 3.99VMRR147 pKa = 11.84EE148 pKa = 3.94IMTHH152 pKa = 4.33ATSRR156 pKa = 11.84EE157 pKa = 4.06EE158 pKa = 3.93YY159 pKa = 10.39LSLVQSSLPYY169 pKa = 10.68DD170 pKa = 3.27WATKK174 pKa = 10.48LNYY177 pKa = 9.62FEE179 pKa = 5.23YY180 pKa = 10.2SASRR184 pKa = 11.84LFPDD188 pKa = 3.2IAEE191 pKa = 5.11PYY193 pKa = 7.99TNPHH197 pKa = 6.38PTTEE201 pKa = 4.23YY202 pKa = 10.94DD203 pKa = 3.4LHH205 pKa = 7.92CNEE208 pKa = 5.52TIEE211 pKa = 5.55DD212 pKa = 3.55WLKK215 pKa = 10.05PNIYY219 pKa = 9.58QVSPQAYY226 pKa = 8.42KK227 pKa = 10.88LLEE230 pKa = 4.21PSCLSLEE237 pKa = 4.11QAIADD242 pKa = 4.68LEE244 pKa = 4.34WLDD247 pKa = 3.51DD248 pKa = 3.85TTRR251 pKa = 11.84MLQEE255 pKa = 4.21KK256 pKa = 9.35EE257 pKa = 4.3RR258 pKa = 11.84EE259 pKa = 4.18ASTSAAQHH267 pKa = 5.69GQVKK271 pKa = 10.29HH272 pKa = 6.52PGLEE276 pKa = 4.15ASDD279 pKa = 3.77DD280 pKa = 3.82TTTGKK285 pKa = 9.01TISTGLHH292 pKa = 4.59MMKK295 pKa = 10.4KK296 pKa = 10.09LSTMSLTTFPSSSVRR311 pKa = 11.84AGNDD315 pKa = 2.47SSAAKK320 pKa = 8.85KK321 pKa = 6.24TTT323 pKa = 3.14
Molecular weight: 37.03 kDa
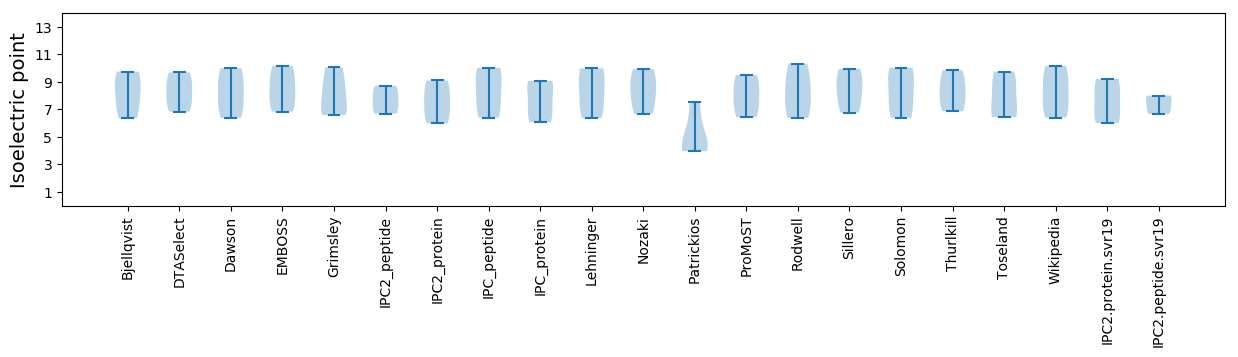
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q00336|MP_PASVK Movement protein OS=Panicum streak virus (isolate Kenya) OX=268780 GN=V2 PE=3 SV=1
MM1 pKa = 7.63SGALKK6 pKa = 10.35RR7 pKa = 11.84KK8 pKa = 10.08RR9 pKa = 11.84SDD11 pKa = 2.87EE12 pKa = 4.37VAWSRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84PVKK22 pKa = 10.48KK23 pKa = 9.39PVRR26 pKa = 11.84RR27 pKa = 11.84APPPRR32 pKa = 11.84AGPSVRR38 pKa = 11.84RR39 pKa = 11.84GLPSLQIQTLVAAGDD54 pKa = 3.71TMITVPSGGICSLIGTYY71 pKa = 10.72ARR73 pKa = 11.84GSGEE77 pKa = 4.23GEE79 pKa = 3.64RR80 pKa = 11.84HH81 pKa = 4.78TNEE84 pKa = 3.48TLTYY88 pKa = 10.08KK89 pKa = 10.59VALDD93 pKa = 3.36YY94 pKa = 11.55HH95 pKa = 6.49FVATAAACKK104 pKa = 9.87YY105 pKa = 9.37SSIGIGVCWLVYY117 pKa = 9.92DD118 pKa = 4.45AQPTGTAPTVQDD130 pKa = 4.2IFPHH134 pKa = 6.56PATLSAFPYY143 pKa = 7.05TWKK146 pKa = 10.5VGRR149 pKa = 11.84EE150 pKa = 3.83VCHH153 pKa = 6.21RR154 pKa = 11.84FVVKK158 pKa = 10.44RR159 pKa = 11.84RR160 pKa = 11.84WCFTMEE166 pKa = 3.97TNGRR170 pKa = 11.84IGSDD174 pKa = 3.38TPPSNVAWPPCKK186 pKa = 9.74KK187 pKa = 10.36DD188 pKa = 3.04IYY190 pKa = 10.04FHH192 pKa = 6.77KK193 pKa = 10.19FCTGLGVKK201 pKa = 7.99TEE203 pKa = 4.15WKK205 pKa = 10.42NVTDD209 pKa = 4.62GKK211 pKa = 10.74DD212 pKa = 3.27GAIKK216 pKa = 10.54KK217 pKa = 9.94GGFYY221 pKa = 10.35IVIAPGNVEE230 pKa = 4.4FTCHH234 pKa = 5.16GQCRR238 pKa = 11.84LYY240 pKa = 10.55FKK242 pKa = 10.94SVGNQQ247 pKa = 2.8
MM1 pKa = 7.63SGALKK6 pKa = 10.35RR7 pKa = 11.84KK8 pKa = 10.08RR9 pKa = 11.84SDD11 pKa = 2.87EE12 pKa = 4.37VAWSRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84PVKK22 pKa = 10.48KK23 pKa = 9.39PVRR26 pKa = 11.84RR27 pKa = 11.84APPPRR32 pKa = 11.84AGPSVRR38 pKa = 11.84RR39 pKa = 11.84GLPSLQIQTLVAAGDD54 pKa = 3.71TMITVPSGGICSLIGTYY71 pKa = 10.72ARR73 pKa = 11.84GSGEE77 pKa = 4.23GEE79 pKa = 3.64RR80 pKa = 11.84HH81 pKa = 4.78TNEE84 pKa = 3.48TLTYY88 pKa = 10.08KK89 pKa = 10.59VALDD93 pKa = 3.36YY94 pKa = 11.55HH95 pKa = 6.49FVATAAACKK104 pKa = 9.87YY105 pKa = 9.37SSIGIGVCWLVYY117 pKa = 9.92DD118 pKa = 4.45AQPTGTAPTVQDD130 pKa = 4.2IFPHH134 pKa = 6.56PATLSAFPYY143 pKa = 7.05TWKK146 pKa = 10.5VGRR149 pKa = 11.84EE150 pKa = 3.83VCHH153 pKa = 6.21RR154 pKa = 11.84FVVKK158 pKa = 10.44RR159 pKa = 11.84RR160 pKa = 11.84WCFTMEE166 pKa = 3.97TNGRR170 pKa = 11.84IGSDD174 pKa = 3.38TPPSNVAWPPCKK186 pKa = 9.74KK187 pKa = 10.36DD188 pKa = 3.04IYY190 pKa = 10.04FHH192 pKa = 6.77KK193 pKa = 10.19FCTGLGVKK201 pKa = 7.99TEE203 pKa = 4.15WKK205 pKa = 10.42NVTDD209 pKa = 4.62GKK211 pKa = 10.74DD212 pKa = 3.27GAIKK216 pKa = 10.54KK217 pKa = 9.94GGFYY221 pKa = 10.35IVIAPGNVEE230 pKa = 4.4FTCHH234 pKa = 5.16GQCRR238 pKa = 11.84LYY240 pKa = 10.55FKK242 pKa = 10.94SVGNQQ247 pKa = 2.8
Molecular weight: 27.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1037 |
108 |
359 |
259.3 |
29.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.172 ± 0.662 | 2.7 ± 0.298 |
5.207 ± 0.412 | 6.75 ± 1.225 |
3.279 ± 0.212 | 5.786 ± 1.172 |
2.989 ± 0.56 | 4.725 ± 0.349 |
6.847 ± 0.707 | 7.04 ± 0.837 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.446 ± 0.273 | 2.797 ± 0.557 |
7.425 ± 0.94 | 2.604 ± 0.07 |
6.75 ± 0.539 | 7.522 ± 0.699 |
7.618 ± 1.111 | 5.4 ± 1.216 |
2.122 ± 0.224 | 4.822 ± 0.639 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
