
French bean severe leaf curl virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Capulavirus
Average proteome isoelectric point is 7.61
Get precalculated fractions of proteins
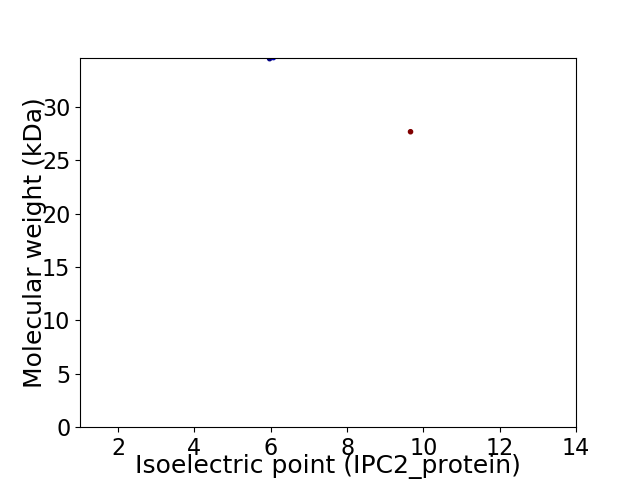
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J7G1P2|J7G1P2_9GEMI Capsid protein OS=French bean severe leaf curl virus OX=1218727 GN=CP PE=3 SV=1
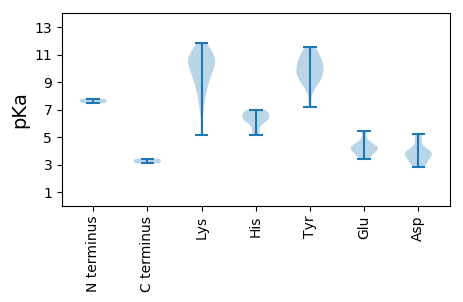
MM1 pKa = 7.78PRR3 pKa = 11.84RR4 pKa = 11.84NNNSFRR10 pKa = 11.84LQGKK14 pKa = 9.58SIFLTYY20 pKa = 9.11PKK22 pKa = 10.58CPLTPLFVIDD32 pKa = 4.87YY33 pKa = 9.9LYY35 pKa = 11.05QLLKK39 pKa = 10.77NFNPIYY45 pKa = 10.86ARR47 pKa = 11.84VCTEE51 pKa = 3.42NHH53 pKa = 6.07QDD55 pKa = 3.87GEE57 pKa = 4.4PHH59 pKa = 6.43LHH61 pKa = 6.94CLVQLDD67 pKa = 3.87KK68 pKa = 11.31RR69 pKa = 11.84FNTTSQRR76 pKa = 11.84YY77 pKa = 8.69FDD79 pKa = 3.87ISDD82 pKa = 3.71PNRR85 pKa = 11.84TGVYY89 pKa = 9.84HH90 pKa = 6.69PNCQVPRR97 pKa = 11.84RR98 pKa = 11.84DD99 pKa = 3.84ADD101 pKa = 3.58VADD104 pKa = 4.81YY105 pKa = 10.27IAKK108 pKa = 10.16GGQFEE113 pKa = 4.22EE114 pKa = 4.62RR115 pKa = 11.84GILRR119 pKa = 11.84ASRR122 pKa = 11.84RR123 pKa = 11.84SPKK126 pKa = 9.94KK127 pKa = 10.16SRR129 pKa = 11.84DD130 pKa = 3.99SIWTNIINEE139 pKa = 4.36STSKK143 pKa = 10.98SEE145 pKa = 3.65FLGRR149 pKa = 11.84VQIEE153 pKa = 4.17QPYY156 pKa = 9.38VWATQLRR163 pKa = 11.84NLEE166 pKa = 4.0YY167 pKa = 10.4AANSKK172 pKa = 9.6WPEE175 pKa = 3.88QPSVYY180 pKa = 9.68IPKK183 pKa = 8.49WTVFNNVPEE192 pKa = 5.28PIRR195 pKa = 11.84EE196 pKa = 4.03WADD199 pKa = 3.31TNLFTVSLQSMQLVQPEE216 pKa = 3.99ISVTDD221 pKa = 3.68MQWAHH226 pKa = 6.35NLTEE230 pKa = 5.42DD231 pKa = 5.21FITDD235 pKa = 3.15EE236 pKa = 4.47WIGNSEE242 pKa = 4.27DD243 pKa = 3.75PVVQSFSGSEE253 pKa = 3.82TGPPSDD259 pKa = 5.17SNNCLAQQKK268 pKa = 7.36QAKK271 pKa = 7.39QHH273 pKa = 6.35GPGAWVYY280 pKa = 10.28IIIFVGVLILVCGIILLLTPSS301 pKa = 3.43
MM1 pKa = 7.78PRR3 pKa = 11.84RR4 pKa = 11.84NNNSFRR10 pKa = 11.84LQGKK14 pKa = 9.58SIFLTYY20 pKa = 9.11PKK22 pKa = 10.58CPLTPLFVIDD32 pKa = 4.87YY33 pKa = 9.9LYY35 pKa = 11.05QLLKK39 pKa = 10.77NFNPIYY45 pKa = 10.86ARR47 pKa = 11.84VCTEE51 pKa = 3.42NHH53 pKa = 6.07QDD55 pKa = 3.87GEE57 pKa = 4.4PHH59 pKa = 6.43LHH61 pKa = 6.94CLVQLDD67 pKa = 3.87KK68 pKa = 11.31RR69 pKa = 11.84FNTTSQRR76 pKa = 11.84YY77 pKa = 8.69FDD79 pKa = 3.87ISDD82 pKa = 3.71PNRR85 pKa = 11.84TGVYY89 pKa = 9.84HH90 pKa = 6.69PNCQVPRR97 pKa = 11.84RR98 pKa = 11.84DD99 pKa = 3.84ADD101 pKa = 3.58VADD104 pKa = 4.81YY105 pKa = 10.27IAKK108 pKa = 10.16GGQFEE113 pKa = 4.22EE114 pKa = 4.62RR115 pKa = 11.84GILRR119 pKa = 11.84ASRR122 pKa = 11.84RR123 pKa = 11.84SPKK126 pKa = 9.94KK127 pKa = 10.16SRR129 pKa = 11.84DD130 pKa = 3.99SIWTNIINEE139 pKa = 4.36STSKK143 pKa = 10.98SEE145 pKa = 3.65FLGRR149 pKa = 11.84VQIEE153 pKa = 4.17QPYY156 pKa = 9.38VWATQLRR163 pKa = 11.84NLEE166 pKa = 4.0YY167 pKa = 10.4AANSKK172 pKa = 9.6WPEE175 pKa = 3.88QPSVYY180 pKa = 9.68IPKK183 pKa = 8.49WTVFNNVPEE192 pKa = 5.28PIRR195 pKa = 11.84EE196 pKa = 4.03WADD199 pKa = 3.31TNLFTVSLQSMQLVQPEE216 pKa = 3.99ISVTDD221 pKa = 3.68MQWAHH226 pKa = 6.35NLTEE230 pKa = 5.42DD231 pKa = 5.21FITDD235 pKa = 3.15EE236 pKa = 4.47WIGNSEE242 pKa = 4.27DD243 pKa = 3.75PVVQSFSGSEE253 pKa = 3.82TGPPSDD259 pKa = 5.17SNNCLAQQKK268 pKa = 7.36QAKK271 pKa = 7.39QHH273 pKa = 6.35GPGAWVYY280 pKa = 10.28IIIFVGVLILVCGIILLLTPSS301 pKa = 3.43
Molecular weight: 34.54 kDa
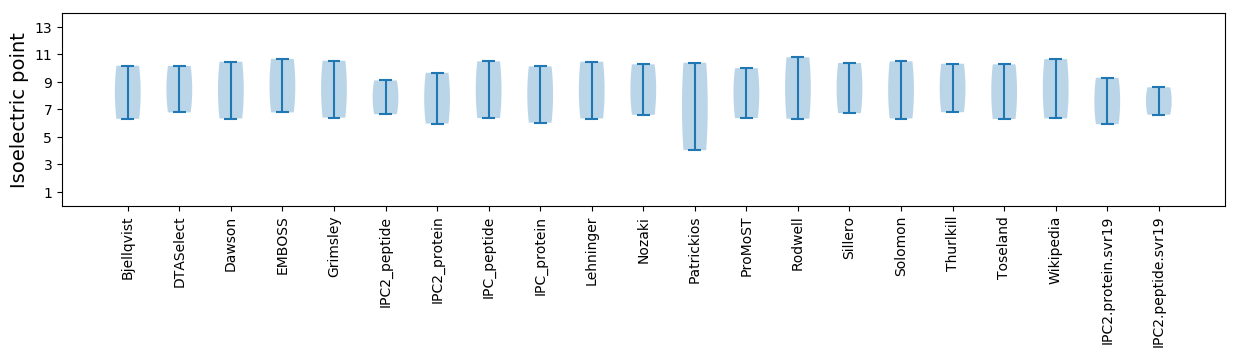
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J7G1P2|J7G1P2_9GEMI Capsid protein OS=French bean severe leaf curl virus OX=1218727 GN=CP PE=3 SV=1
MM1 pKa = 7.47ARR3 pKa = 11.84TRR5 pKa = 11.84SGRR8 pKa = 11.84QYY10 pKa = 10.92SAQAPSWGRR19 pKa = 11.84KK20 pKa = 5.17KK21 pKa = 9.17TRR23 pKa = 11.84TPRR26 pKa = 11.84SRR28 pKa = 11.84PTLIGPIRR36 pKa = 11.84RR37 pKa = 11.84PNYY40 pKa = 9.4QIKK43 pKa = 8.5TRR45 pKa = 11.84YY46 pKa = 9.14APHH49 pKa = 6.95RR50 pKa = 11.84PQTKK54 pKa = 8.93IHH56 pKa = 6.21SLSAARR62 pKa = 11.84VVSGSDD68 pKa = 2.89NGYY71 pKa = 8.75GWHH74 pKa = 6.87ISDD77 pKa = 4.11VPIGSGFEE85 pKa = 4.22DD86 pKa = 3.08RR87 pKa = 11.84HH88 pKa = 6.35SDD90 pKa = 4.03KK91 pKa = 10.91IRR93 pKa = 11.84ILNFNFKK100 pKa = 10.56LQMRR104 pKa = 11.84TSQQGQNTSCWHH116 pKa = 5.12NVYY119 pKa = 10.72LFLVRR124 pKa = 11.84DD125 pKa = 3.94NSGGTAVPKK134 pKa = 10.51FNSICMMDD142 pKa = 3.69NSNPSTAEE150 pKa = 3.47IDD152 pKa = 3.61HH153 pKa = 6.87DD154 pKa = 4.33SKK156 pKa = 11.82DD157 pKa = 2.99RR158 pKa = 11.84FTIMRR163 pKa = 11.84RR164 pKa = 11.84WRR166 pKa = 11.84FQFKK170 pKa = 10.73GNSTSGSVAYY180 pKa = 10.25DD181 pKa = 3.62CARR184 pKa = 11.84NTYY187 pKa = 9.93DD188 pKa = 2.84FRR190 pKa = 11.84KK191 pKa = 9.1FVKK194 pKa = 10.27LSSITEE200 pKa = 4.36FKK202 pKa = 10.83SATSGSYY209 pKa = 11.56ANTQKK214 pKa = 10.86NAWVLYY220 pKa = 7.21FVPQTYY226 pKa = 11.35DD227 pKa = 3.06MTVDD231 pKa = 3.31GHH233 pKa = 5.43CTIKK237 pKa = 10.77YY238 pKa = 9.39VSIVV242 pKa = 3.08
MM1 pKa = 7.47ARR3 pKa = 11.84TRR5 pKa = 11.84SGRR8 pKa = 11.84QYY10 pKa = 10.92SAQAPSWGRR19 pKa = 11.84KK20 pKa = 5.17KK21 pKa = 9.17TRR23 pKa = 11.84TPRR26 pKa = 11.84SRR28 pKa = 11.84PTLIGPIRR36 pKa = 11.84RR37 pKa = 11.84PNYY40 pKa = 9.4QIKK43 pKa = 8.5TRR45 pKa = 11.84YY46 pKa = 9.14APHH49 pKa = 6.95RR50 pKa = 11.84PQTKK54 pKa = 8.93IHH56 pKa = 6.21SLSAARR62 pKa = 11.84VVSGSDD68 pKa = 2.89NGYY71 pKa = 8.75GWHH74 pKa = 6.87ISDD77 pKa = 4.11VPIGSGFEE85 pKa = 4.22DD86 pKa = 3.08RR87 pKa = 11.84HH88 pKa = 6.35SDD90 pKa = 4.03KK91 pKa = 10.91IRR93 pKa = 11.84ILNFNFKK100 pKa = 10.56LQMRR104 pKa = 11.84TSQQGQNTSCWHH116 pKa = 5.12NVYY119 pKa = 10.72LFLVRR124 pKa = 11.84DD125 pKa = 3.94NSGGTAVPKK134 pKa = 10.51FNSICMMDD142 pKa = 3.69NSNPSTAEE150 pKa = 3.47IDD152 pKa = 3.61HH153 pKa = 6.87DD154 pKa = 4.33SKK156 pKa = 11.82DD157 pKa = 2.99RR158 pKa = 11.84FTIMRR163 pKa = 11.84RR164 pKa = 11.84WRR166 pKa = 11.84FQFKK170 pKa = 10.73GNSTSGSVAYY180 pKa = 10.25DD181 pKa = 3.62CARR184 pKa = 11.84NTYY187 pKa = 9.93DD188 pKa = 2.84FRR190 pKa = 11.84KK191 pKa = 9.1FVKK194 pKa = 10.27LSSITEE200 pKa = 4.36FKK202 pKa = 10.83SATSGSYY209 pKa = 11.56ANTQKK214 pKa = 10.86NAWVLYY220 pKa = 7.21FVPQTYY226 pKa = 11.35DD227 pKa = 3.06MTVDD231 pKa = 3.31GHH233 pKa = 5.43CTIKK237 pKa = 10.77YY238 pKa = 9.39VSIVV242 pKa = 3.08
Molecular weight: 27.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
543 |
242 |
301 |
271.5 |
31.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.788 ± 0.382 | 1.842 ± 0.123 |
5.157 ± 0.141 | 3.499 ± 1.478 |
4.604 ± 0.232 | 5.341 ± 0.561 |
2.394 ± 0.326 | 6.446 ± 0.432 |
4.788 ± 0.652 | 5.893 ± 1.692 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.657 ± 0.537 | 6.262 ± 0.312 |
6.077 ± 1.002 | 5.709 ± 0.761 |
7.182 ± 1.248 | 9.208 ± 1.274 |
6.63 ± 0.799 | 6.077 ± 0.461 |
2.394 ± 0.214 | 4.052 ± 0.323 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
