
Zobellia galactanivorans (strain DSM 12802 / CCUG 47099 / CIP 106680 / NCIMB 13871 / Dsij)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia;
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
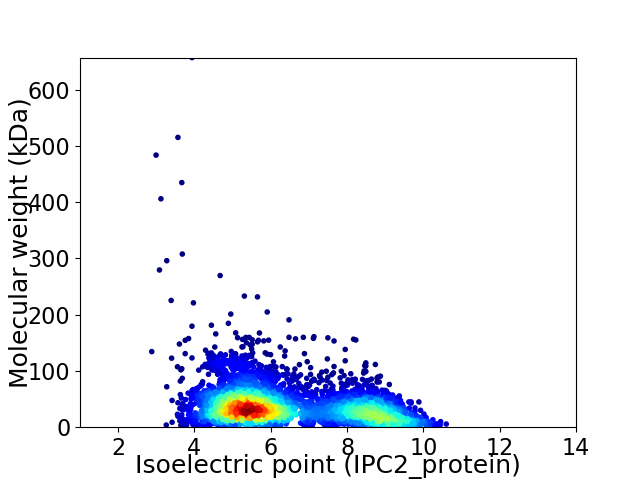
Virtual 2D-PAGE plot for 4708 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G0LC95|G0LC95_ZOBGA Uncharacterized protein OS=Zobellia galactanivorans (strain DSM 12802 / CCUG 47099 / CIP 106680 / NCIMB 13871 / Dsij) OX=63186 GN=zobellia_2605 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 10.41GFKK5 pKa = 10.45YY6 pKa = 10.88YY7 pKa = 10.14IGTITIFSIMTCSLISCSNDD27 pKa = 3.35DD28 pKa = 5.88DD29 pKa = 5.36DD30 pKa = 7.22DD31 pKa = 4.8EE32 pKa = 5.41YY33 pKa = 11.55LGNWVGRR40 pKa = 11.84SVFDD44 pKa = 3.46GSPRR48 pKa = 11.84SGTSSFTIGNMGYY61 pKa = 8.46TGVGYY66 pKa = 10.68DD67 pKa = 3.57GDD69 pKa = 5.04DD70 pKa = 3.62YY71 pKa = 11.4LTSFWSYY78 pKa = 12.15DD79 pKa = 3.04MDD81 pKa = 5.72GDD83 pKa = 4.27FWSQKK88 pKa = 10.74ADD90 pKa = 4.28FIGSARR96 pKa = 11.84NAAVGFEE103 pKa = 4.14VDD105 pKa = 3.08GKK107 pKa = 11.09GYY109 pKa = 10.2IGSGYY114 pKa = 10.68DD115 pKa = 5.01GLDD118 pKa = 3.48EE119 pKa = 6.17LNDD122 pKa = 4.39FYY124 pKa = 11.59AYY126 pKa = 10.39DD127 pKa = 4.02PSSNSWQEE135 pKa = 3.81IASLPSTGRR144 pKa = 11.84RR145 pKa = 11.84SAVAFGMNGFGYY157 pKa = 10.39FGTGFDD163 pKa = 4.34GDD165 pKa = 3.78NDD167 pKa = 3.98RR168 pKa = 11.84KK169 pKa = 10.91DD170 pKa = 3.11FWKK173 pKa = 10.64YY174 pKa = 11.1DD175 pKa = 3.55PSTDD179 pKa = 2.98SWSEE183 pKa = 3.65LVGFGGDD190 pKa = 3.25KK191 pKa = 10.27RR192 pKa = 11.84RR193 pKa = 11.84SATTFTIGDD202 pKa = 3.7KK203 pKa = 10.82VYY205 pKa = 11.06LGTGVSNGIYY215 pKa = 10.32LDD217 pKa = 4.47DD218 pKa = 4.53FWAFDD223 pKa = 3.73ATSEE227 pKa = 4.09TWTKK231 pKa = 11.12KK232 pKa = 10.48LDD234 pKa = 3.73LDD236 pKa = 4.33EE237 pKa = 5.31EE238 pKa = 4.88DD239 pKa = 5.06DD240 pKa = 4.18YY241 pKa = 11.99SITRR245 pKa = 11.84SNAVGFTLNGYY256 pKa = 10.14GYY258 pKa = 8.96IACGTSSGTLASVWQYY274 pKa = 11.92DD275 pKa = 3.53PSTDD279 pKa = 3.1EE280 pKa = 4.53WEE282 pKa = 4.3LKK284 pKa = 9.96TDD286 pKa = 4.07FEE288 pKa = 4.65GTSRR292 pKa = 11.84QDD294 pKa = 2.87AVVFSNGSKK303 pKa = 10.5AVVGLGRR310 pKa = 11.84TGSLYY315 pKa = 10.9LDD317 pKa = 4.14DD318 pKa = 5.85LYY320 pKa = 11.51EE321 pKa = 4.15FFPFDD326 pKa = 4.08EE327 pKa = 4.99YY328 pKa = 11.59DD329 pKa = 4.75DD330 pKa = 4.28EE331 pKa = 4.99DD332 pKa = 3.69
MM1 pKa = 7.67KK2 pKa = 10.41GFKK5 pKa = 10.45YY6 pKa = 10.88YY7 pKa = 10.14IGTITIFSIMTCSLISCSNDD27 pKa = 3.35DD28 pKa = 5.88DD29 pKa = 5.36DD30 pKa = 7.22DD31 pKa = 4.8EE32 pKa = 5.41YY33 pKa = 11.55LGNWVGRR40 pKa = 11.84SVFDD44 pKa = 3.46GSPRR48 pKa = 11.84SGTSSFTIGNMGYY61 pKa = 8.46TGVGYY66 pKa = 10.68DD67 pKa = 3.57GDD69 pKa = 5.04DD70 pKa = 3.62YY71 pKa = 11.4LTSFWSYY78 pKa = 12.15DD79 pKa = 3.04MDD81 pKa = 5.72GDD83 pKa = 4.27FWSQKK88 pKa = 10.74ADD90 pKa = 4.28FIGSARR96 pKa = 11.84NAAVGFEE103 pKa = 4.14VDD105 pKa = 3.08GKK107 pKa = 11.09GYY109 pKa = 10.2IGSGYY114 pKa = 10.68DD115 pKa = 5.01GLDD118 pKa = 3.48EE119 pKa = 6.17LNDD122 pKa = 4.39FYY124 pKa = 11.59AYY126 pKa = 10.39DD127 pKa = 4.02PSSNSWQEE135 pKa = 3.81IASLPSTGRR144 pKa = 11.84RR145 pKa = 11.84SAVAFGMNGFGYY157 pKa = 10.39FGTGFDD163 pKa = 4.34GDD165 pKa = 3.78NDD167 pKa = 3.98RR168 pKa = 11.84KK169 pKa = 10.91DD170 pKa = 3.11FWKK173 pKa = 10.64YY174 pKa = 11.1DD175 pKa = 3.55PSTDD179 pKa = 2.98SWSEE183 pKa = 3.65LVGFGGDD190 pKa = 3.25KK191 pKa = 10.27RR192 pKa = 11.84RR193 pKa = 11.84SATTFTIGDD202 pKa = 3.7KK203 pKa = 10.82VYY205 pKa = 11.06LGTGVSNGIYY215 pKa = 10.32LDD217 pKa = 4.47DD218 pKa = 4.53FWAFDD223 pKa = 3.73ATSEE227 pKa = 4.09TWTKK231 pKa = 11.12KK232 pKa = 10.48LDD234 pKa = 3.73LDD236 pKa = 4.33EE237 pKa = 5.31EE238 pKa = 4.88DD239 pKa = 5.06DD240 pKa = 4.18YY241 pKa = 11.99SITRR245 pKa = 11.84SNAVGFTLNGYY256 pKa = 10.14GYY258 pKa = 8.96IACGTSSGTLASVWQYY274 pKa = 11.92DD275 pKa = 3.53PSTDD279 pKa = 3.1EE280 pKa = 4.53WEE282 pKa = 4.3LKK284 pKa = 9.96TDD286 pKa = 4.07FEE288 pKa = 4.65GTSRR292 pKa = 11.84QDD294 pKa = 2.87AVVFSNGSKK303 pKa = 10.5AVVGLGRR310 pKa = 11.84TGSLYY315 pKa = 10.9LDD317 pKa = 4.14DD318 pKa = 5.85LYY320 pKa = 11.51EE321 pKa = 4.15FFPFDD326 pKa = 4.08EE327 pKa = 4.99YY328 pKa = 11.59DD329 pKa = 4.75DD330 pKa = 4.28EE331 pKa = 4.99DD332 pKa = 3.69
Molecular weight: 36.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G0L0S1|G0L0S1_ZOBGA GntR-type transcriptional regulator OS=Zobellia galactanivorans (strain DSM 12802 / CCUG 47099 / CIP 106680 / NCIMB 13871 / Dsij) OX=63186 GN=zobellia_329 PE=4 SV=1
MM1 pKa = 7.47NKK3 pKa = 9.56NLGYY7 pKa = 8.81TGYY10 pKa = 9.09WAEE13 pKa = 4.2RR14 pKa = 11.84RR15 pKa = 11.84FGFDD19 pKa = 2.25KK20 pKa = 10.71CRR22 pKa = 11.84YY23 pKa = 8.34KK24 pKa = 10.47VQEE27 pKa = 4.52GIFEE31 pKa = 4.22IFRR34 pKa = 11.84AIRR37 pKa = 11.84GVFGMLVRR45 pKa = 11.84RR46 pKa = 11.84FALISGFQPNLISLIKK62 pKa = 10.51LL63 pKa = 3.44
MM1 pKa = 7.47NKK3 pKa = 9.56NLGYY7 pKa = 8.81TGYY10 pKa = 9.09WAEE13 pKa = 4.2RR14 pKa = 11.84RR15 pKa = 11.84FGFDD19 pKa = 2.25KK20 pKa = 10.71CRR22 pKa = 11.84YY23 pKa = 8.34KK24 pKa = 10.47VQEE27 pKa = 4.52GIFEE31 pKa = 4.22IFRR34 pKa = 11.84AIRR37 pKa = 11.84GVFGMLVRR45 pKa = 11.84RR46 pKa = 11.84FALISGFQPNLISLIKK62 pKa = 10.51LL63 pKa = 3.44
Molecular weight: 7.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1661998 |
25 |
6203 |
353.0 |
39.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.675 ± 0.029 | 0.739 ± 0.011 |
6.01 ± 0.054 | 6.659 ± 0.033 |
5.028 ± 0.025 | 7.096 ± 0.035 |
1.853 ± 0.021 | 6.995 ± 0.031 |
7.219 ± 0.053 | 9.233 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.22 ± 0.02 | 5.605 ± 0.036 |
3.74 ± 0.024 | 3.272 ± 0.018 |
3.754 ± 0.024 | 6.501 ± 0.029 |
5.77 ± 0.046 | 6.327 ± 0.029 |
1.197 ± 0.013 | 4.107 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
