
Hudisavirus sp.
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
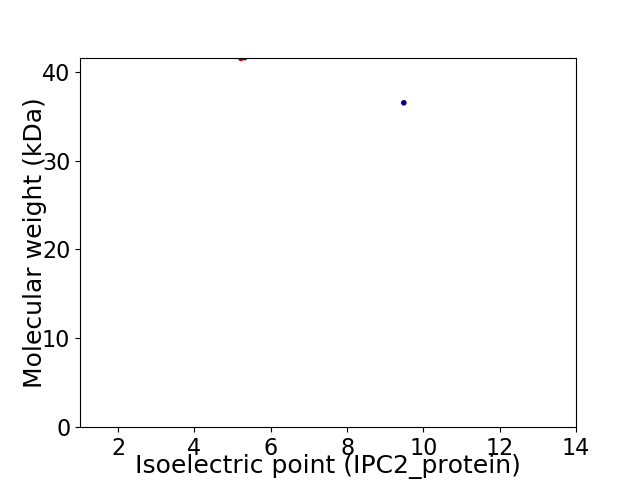
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A223Q5V2|A0A223Q5V2_9VIRU Putative capsid protein OS=Hudisavirus sp. OX=2021738 PE=4 SV=1
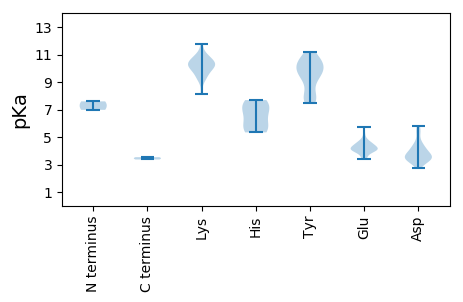
MM1 pKa = 7.59IDD3 pKa = 3.06SMAAEE8 pKa = 4.28TAAKK12 pKa = 9.75RR13 pKa = 11.84WCFTINNPTDD23 pKa = 3.38EE24 pKa = 4.5DD25 pKa = 4.29KK26 pKa = 10.99FWEE29 pKa = 4.16NAEE32 pKa = 4.0QQEE35 pKa = 4.1QLEE38 pKa = 4.27YY39 pKa = 10.87LIVQHH44 pKa = 6.33EE45 pKa = 4.58VGEE48 pKa = 4.32QGTPHH53 pKa = 5.68YY54 pKa = 10.51QGFLILKK61 pKa = 9.3RR62 pKa = 11.84KK63 pKa = 9.22NRR65 pKa = 11.84LSWLKK70 pKa = 10.97KK71 pKa = 9.19NINSRR76 pKa = 11.84AHH78 pKa = 5.34WEE80 pKa = 4.11KK81 pKa = 10.26TRR83 pKa = 11.84GTDD86 pKa = 3.35QQAADD91 pKa = 4.0YY92 pKa = 10.33CRR94 pKa = 11.84KK95 pKa = 10.26DD96 pKa = 3.38DD97 pKa = 3.67THH99 pKa = 7.69PEE101 pKa = 3.64GSLRR105 pKa = 11.84FEE107 pKa = 3.93WGTLKK112 pKa = 9.92TALKK116 pKa = 10.53RR117 pKa = 11.84KK118 pKa = 10.06DD119 pKa = 3.57RR120 pKa = 11.84DD121 pKa = 3.77CLEE124 pKa = 3.95EE125 pKa = 4.02AVIEE129 pKa = 4.16EE130 pKa = 4.24VKK132 pKa = 10.58KK133 pKa = 10.91IQDD136 pKa = 3.11EE137 pKa = 4.61GFRR140 pKa = 11.84SVRR143 pKa = 11.84DD144 pKa = 3.14IDD146 pKa = 3.95PQVLARR152 pKa = 11.84PGFMAAYY159 pKa = 8.95NALTADD165 pKa = 3.53MLGVYY170 pKa = 9.74RR171 pKa = 11.84PEE173 pKa = 3.99LKK175 pKa = 9.86IVTMVGPPGTGKK187 pKa = 10.42SYY189 pKa = 10.9AINTLFPHH197 pKa = 7.24AGRR200 pKa = 11.84AIMGNGGTWFANPTATVMVFEE221 pKa = 4.84EE222 pKa = 4.5FAGQIPLQKK231 pKa = 9.48MLKK234 pKa = 10.37LLDD237 pKa = 4.66PYY239 pKa = 11.18PMALEE244 pKa = 4.52VKK246 pKa = 10.15GGMRR250 pKa = 11.84PAMYY254 pKa = 9.13TLVIITSNTRR264 pKa = 11.84PDD266 pKa = 2.75GWYY269 pKa = 10.16KK270 pKa = 10.8EE271 pKa = 4.2EE272 pKa = 4.01EE273 pKa = 4.14QGGKK277 pKa = 8.16RR278 pKa = 11.84TDD280 pKa = 3.27ALLALWDD287 pKa = 3.9RR288 pKa = 11.84LGFSNGTNRR297 pKa = 11.84CYY299 pKa = 9.72RR300 pKa = 11.84TCGTYY305 pKa = 10.46LEE307 pKa = 4.79PVRR310 pKa = 11.84GMALGAAGTWIDD322 pKa = 3.61NTRR325 pKa = 11.84TWFMNEE331 pKa = 3.48LAKK334 pKa = 10.47AAHH337 pKa = 5.57VEE339 pKa = 4.09EE340 pKa = 5.71HH341 pKa = 6.84EE342 pKa = 4.38EE343 pKa = 4.07LSDD346 pKa = 3.54EE347 pKa = 4.47ALSQVEE353 pKa = 3.96QDD355 pKa = 3.96KK356 pKa = 11.74LEE358 pKa = 4.57DD359 pKa = 5.55DD360 pKa = 3.78IASLDD365 pKa = 3.59VV366 pKa = 3.54
MM1 pKa = 7.59IDD3 pKa = 3.06SMAAEE8 pKa = 4.28TAAKK12 pKa = 9.75RR13 pKa = 11.84WCFTINNPTDD23 pKa = 3.38EE24 pKa = 4.5DD25 pKa = 4.29KK26 pKa = 10.99FWEE29 pKa = 4.16NAEE32 pKa = 4.0QQEE35 pKa = 4.1QLEE38 pKa = 4.27YY39 pKa = 10.87LIVQHH44 pKa = 6.33EE45 pKa = 4.58VGEE48 pKa = 4.32QGTPHH53 pKa = 5.68YY54 pKa = 10.51QGFLILKK61 pKa = 9.3RR62 pKa = 11.84KK63 pKa = 9.22NRR65 pKa = 11.84LSWLKK70 pKa = 10.97KK71 pKa = 9.19NINSRR76 pKa = 11.84AHH78 pKa = 5.34WEE80 pKa = 4.11KK81 pKa = 10.26TRR83 pKa = 11.84GTDD86 pKa = 3.35QQAADD91 pKa = 4.0YY92 pKa = 10.33CRR94 pKa = 11.84KK95 pKa = 10.26DD96 pKa = 3.38DD97 pKa = 3.67THH99 pKa = 7.69PEE101 pKa = 3.64GSLRR105 pKa = 11.84FEE107 pKa = 3.93WGTLKK112 pKa = 9.92TALKK116 pKa = 10.53RR117 pKa = 11.84KK118 pKa = 10.06DD119 pKa = 3.57RR120 pKa = 11.84DD121 pKa = 3.77CLEE124 pKa = 3.95EE125 pKa = 4.02AVIEE129 pKa = 4.16EE130 pKa = 4.24VKK132 pKa = 10.58KK133 pKa = 10.91IQDD136 pKa = 3.11EE137 pKa = 4.61GFRR140 pKa = 11.84SVRR143 pKa = 11.84DD144 pKa = 3.14IDD146 pKa = 3.95PQVLARR152 pKa = 11.84PGFMAAYY159 pKa = 8.95NALTADD165 pKa = 3.53MLGVYY170 pKa = 9.74RR171 pKa = 11.84PEE173 pKa = 3.99LKK175 pKa = 9.86IVTMVGPPGTGKK187 pKa = 10.42SYY189 pKa = 10.9AINTLFPHH197 pKa = 7.24AGRR200 pKa = 11.84AIMGNGGTWFANPTATVMVFEE221 pKa = 4.84EE222 pKa = 4.5FAGQIPLQKK231 pKa = 9.48MLKK234 pKa = 10.37LLDD237 pKa = 4.66PYY239 pKa = 11.18PMALEE244 pKa = 4.52VKK246 pKa = 10.15GGMRR250 pKa = 11.84PAMYY254 pKa = 9.13TLVIITSNTRR264 pKa = 11.84PDD266 pKa = 2.75GWYY269 pKa = 10.16KK270 pKa = 10.8EE271 pKa = 4.2EE272 pKa = 4.01EE273 pKa = 4.14QGGKK277 pKa = 8.16RR278 pKa = 11.84TDD280 pKa = 3.27ALLALWDD287 pKa = 3.9RR288 pKa = 11.84LGFSNGTNRR297 pKa = 11.84CYY299 pKa = 9.72RR300 pKa = 11.84TCGTYY305 pKa = 10.46LEE307 pKa = 4.79PVRR310 pKa = 11.84GMALGAAGTWIDD322 pKa = 3.61NTRR325 pKa = 11.84TWFMNEE331 pKa = 3.48LAKK334 pKa = 10.47AAHH337 pKa = 5.57VEE339 pKa = 4.09EE340 pKa = 5.71HH341 pKa = 6.84EE342 pKa = 4.38EE343 pKa = 4.07LSDD346 pKa = 3.54EE347 pKa = 4.47ALSQVEE353 pKa = 3.96QDD355 pKa = 3.96KK356 pKa = 11.74LEE358 pKa = 4.57DD359 pKa = 5.55DD360 pKa = 3.78IASLDD365 pKa = 3.59VV366 pKa = 3.54
Molecular weight: 41.49 kDa
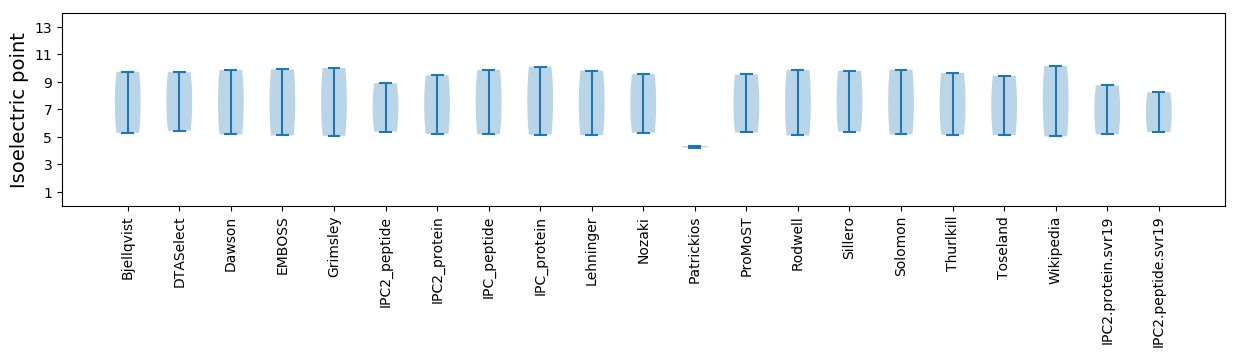
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A223Q5V2|A0A223Q5V2_9VIRU Putative capsid protein OS=Hudisavirus sp. OX=2021738 PE=4 SV=1
MM1 pKa = 6.98PTYY4 pKa = 9.82VYY6 pKa = 10.43KK7 pKa = 10.49YY8 pKa = 7.97RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 7.5GRR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 8.92SRR18 pKa = 11.84YY19 pKa = 8.02RR20 pKa = 11.84RR21 pKa = 11.84YY22 pKa = 9.69SGAYY26 pKa = 7.57SRR28 pKa = 11.84YY29 pKa = 7.74RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84SGTSSTASSRR43 pKa = 11.84GRR45 pKa = 11.84IRR47 pKa = 11.84VRR49 pKa = 11.84VPIQQVVTMTVPAGSIDD66 pKa = 4.21SNLLTSSPFYY76 pKa = 7.79WTSSITSAPYY86 pKa = 9.63GRR88 pKa = 11.84CGAVATPLYY97 pKa = 10.09RR98 pKa = 11.84AYY100 pKa = 9.82TSLYY104 pKa = 10.57DD105 pKa = 3.56QVKK108 pKa = 10.25CDD110 pKa = 3.55GVVTNLSVVSPIGGGSGAAASALQVIVAYY139 pKa = 10.23DD140 pKa = 3.32RR141 pKa = 11.84QGNFNEE147 pKa = 4.17VLNGDD152 pKa = 4.3GVTVAQLFNFSSAVVRR168 pKa = 11.84SAINNSVAKK177 pKa = 9.78MSRR180 pKa = 11.84SCWASDD186 pKa = 3.31LQEE189 pKa = 3.39RR190 pKa = 11.84TMFHH194 pKa = 7.39DD195 pKa = 4.18CTVSASGNPVNTITDD210 pKa = 3.46GDD212 pKa = 4.02FSSNQQKK219 pKa = 10.31VGYY222 pKa = 7.91FAPLTYY228 pKa = 10.62VGLRR232 pKa = 11.84LAAAAPTSDD241 pKa = 5.37LSIQVLLEE249 pKa = 3.47QTYY252 pKa = 9.51YY253 pKa = 9.61FTFRR257 pKa = 11.84NPKK260 pKa = 10.07FGADD264 pKa = 3.51TSNSASFSVRR274 pKa = 11.84PVTMDD279 pKa = 3.17SAVDD283 pKa = 3.54TRR285 pKa = 11.84TMQSTQVMDD294 pKa = 4.75DD295 pKa = 4.3DD296 pKa = 5.79GGLDD300 pKa = 4.49DD301 pKa = 5.34EE302 pKa = 5.22AAQAAQPAARR312 pKa = 11.84VARR315 pKa = 11.84ATPLSSLFDD324 pKa = 3.59SSIPVPARR332 pKa = 11.84FRR334 pKa = 11.84QQ335 pKa = 3.37
MM1 pKa = 6.98PTYY4 pKa = 9.82VYY6 pKa = 10.43KK7 pKa = 10.49YY8 pKa = 7.97RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 7.5GRR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 8.92SRR18 pKa = 11.84YY19 pKa = 8.02RR20 pKa = 11.84RR21 pKa = 11.84YY22 pKa = 9.69SGAYY26 pKa = 7.57SRR28 pKa = 11.84YY29 pKa = 7.74RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84SGTSSTASSRR43 pKa = 11.84GRR45 pKa = 11.84IRR47 pKa = 11.84VRR49 pKa = 11.84VPIQQVVTMTVPAGSIDD66 pKa = 4.21SNLLTSSPFYY76 pKa = 7.79WTSSITSAPYY86 pKa = 9.63GRR88 pKa = 11.84CGAVATPLYY97 pKa = 10.09RR98 pKa = 11.84AYY100 pKa = 9.82TSLYY104 pKa = 10.57DD105 pKa = 3.56QVKK108 pKa = 10.25CDD110 pKa = 3.55GVVTNLSVVSPIGGGSGAAASALQVIVAYY139 pKa = 10.23DD140 pKa = 3.32RR141 pKa = 11.84QGNFNEE147 pKa = 4.17VLNGDD152 pKa = 4.3GVTVAQLFNFSSAVVRR168 pKa = 11.84SAINNSVAKK177 pKa = 9.78MSRR180 pKa = 11.84SCWASDD186 pKa = 3.31LQEE189 pKa = 3.39RR190 pKa = 11.84TMFHH194 pKa = 7.39DD195 pKa = 4.18CTVSASGNPVNTITDD210 pKa = 3.46GDD212 pKa = 4.02FSSNQQKK219 pKa = 10.31VGYY222 pKa = 7.91FAPLTYY228 pKa = 10.62VGLRR232 pKa = 11.84LAAAAPTSDD241 pKa = 5.37LSIQVLLEE249 pKa = 3.47QTYY252 pKa = 9.51YY253 pKa = 9.61FTFRR257 pKa = 11.84NPKK260 pKa = 10.07FGADD264 pKa = 3.51TSNSASFSVRR274 pKa = 11.84PVTMDD279 pKa = 3.17SAVDD283 pKa = 3.54TRR285 pKa = 11.84TMQSTQVMDD294 pKa = 4.75DD295 pKa = 4.3DD296 pKa = 5.79GGLDD300 pKa = 4.49DD301 pKa = 5.34EE302 pKa = 5.22AAQAAQPAARR312 pKa = 11.84VARR315 pKa = 11.84ATPLSSLFDD324 pKa = 3.59SSIPVPARR332 pKa = 11.84FRR334 pKa = 11.84QQ335 pKa = 3.37
Molecular weight: 36.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
701 |
335 |
366 |
350.5 |
39.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.7 ± 0.512 | 1.284 ± 0.062 |
5.991 ± 0.219 | 5.136 ± 2.698 |
3.566 ± 0.215 | 7.133 ± 0.387 |
1.141 ± 0.577 | 3.709 ± 0.496 |
3.852 ± 1.615 | 7.133 ± 1.204 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.853 ± 0.523 | 3.994 ± 0.078 |
4.565 ± 0.145 | 4.422 ± 0.242 |
7.561 ± 1.159 | 7.703 ± 3.513 |
7.418 ± 0.235 | 6.847 ± 1.647 |
1.712 ± 0.763 | 4.28 ± 0.953 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
