
Lactobacillus phage phiPYB5
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
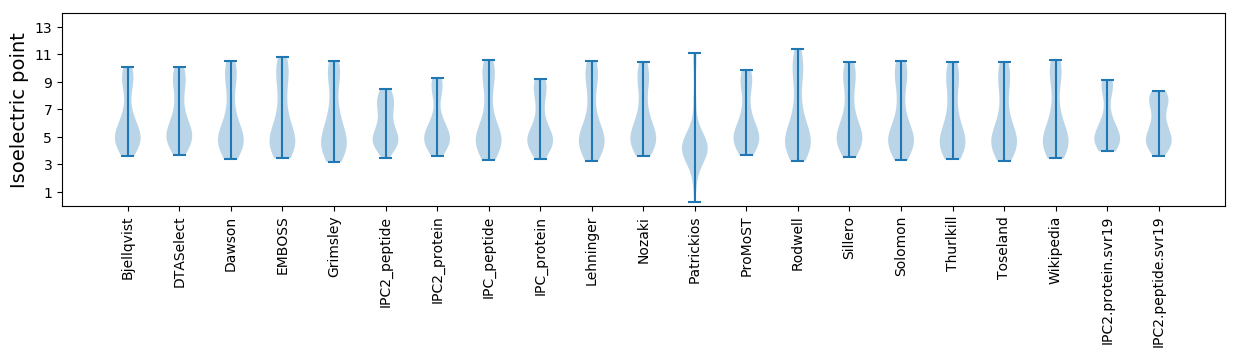
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
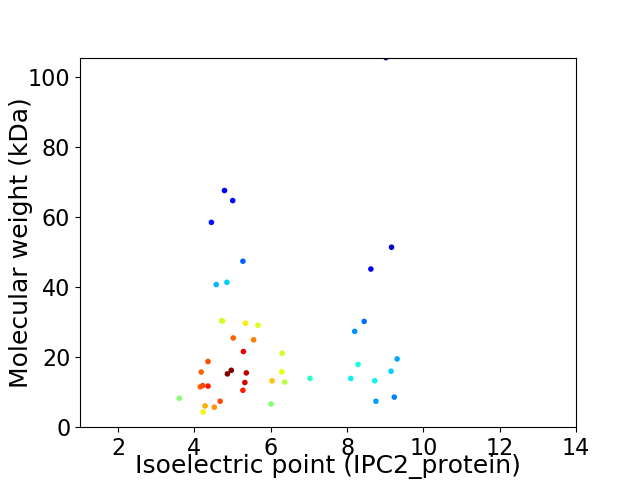
Virtual 2D-PAGE plot for 46 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D2KRC4|D2KRC4_9CAUD BppU_N domain-containing protein OS=Lactobacillus phage phiPYB5 OX=438780 GN=CU5_20 PE=4 SV=1
MM1 pKa = 7.34FNLFGSLEE9 pKa = 4.13MYY11 pKa = 10.68LNNLIKK17 pKa = 10.5VANSYY22 pKa = 10.62GVLDD26 pKa = 3.92QQTGQATTTVILTPPTGLTLDD47 pKa = 3.89SDD49 pKa = 4.19FVDD52 pKa = 5.71AINALWSTIEE62 pKa = 3.92STLNNLQSYY71 pKa = 10.83ANEE74 pKa = 4.1FF75 pKa = 3.5
MM1 pKa = 7.34FNLFGSLEE9 pKa = 4.13MYY11 pKa = 10.68LNNLIKK17 pKa = 10.5VANSYY22 pKa = 10.62GVLDD26 pKa = 3.92QQTGQATTTVILTPPTGLTLDD47 pKa = 3.89SDD49 pKa = 4.19FVDD52 pKa = 5.71AINALWSTIEE62 pKa = 3.92STLNNLQSYY71 pKa = 10.83ANEE74 pKa = 4.1FF75 pKa = 3.5
Molecular weight: 8.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D2KRD3|D2KRD3_9CAUD Uncharacterized protein OS=Lactobacillus phage phiPYB5 OX=438780 GN=CU5_31 PE=4 SV=1
MM1 pKa = 7.96LEE3 pKa = 3.84VCMKK7 pKa = 8.73IAVYY11 pKa = 10.6VRR13 pKa = 11.84VSTLEE18 pKa = 3.83QAEE21 pKa = 4.25SGYY24 pKa = 10.92SIGEE28 pKa = 3.89QTEE31 pKa = 4.12KK32 pKa = 10.08LTAYY36 pKa = 10.28CRR38 pKa = 11.84VKK40 pKa = 10.25DD41 pKa = 3.54WQVAKK46 pKa = 10.39IYY48 pKa = 9.33TDD50 pKa = 3.52PGFSGSSLDD59 pKa = 3.75RR60 pKa = 11.84PAMQQLIGDD69 pKa = 4.39CKK71 pKa = 10.83LGVFDD76 pKa = 5.52AVLVYY81 pKa = 10.81KK82 pKa = 10.59LDD84 pKa = 3.8RR85 pKa = 11.84LSRR88 pKa = 11.84SQKK91 pKa = 8.91DD92 pKa = 3.35TLYY95 pKa = 11.05LIEE98 pKa = 5.4DD99 pKa = 3.6VFNAYY104 pKa = 10.12NIHH107 pKa = 6.31FMSLSEE113 pKa = 4.46NFDD116 pKa = 3.17TSTPFGKK123 pKa = 10.57AMIGLLSVFAQLEE136 pKa = 4.12RR137 pKa = 11.84EE138 pKa = 4.22QIKK141 pKa = 10.5EE142 pKa = 3.7RR143 pKa = 11.84MQMGKK148 pKa = 10.18LGRR151 pKa = 11.84VKK153 pKa = 10.42AGKK156 pKa = 9.81ISAWSNVPFGYY167 pKa = 9.3TKK169 pKa = 10.82VKK171 pKa = 10.57DD172 pKa = 4.02SYY174 pKa = 11.86DD175 pKa = 3.79VDD177 pKa = 3.89PLRR180 pKa = 11.84ADD182 pKa = 3.3IVRR185 pKa = 11.84RR186 pKa = 11.84IYY188 pKa = 11.28ADD190 pKa = 3.39YY191 pKa = 10.85LAGKK195 pKa = 9.92SITKK199 pKa = 9.68IMKK202 pKa = 8.03EE203 pKa = 4.02LNRR206 pKa = 11.84EE207 pKa = 3.74GHH209 pKa = 5.88IGKK212 pKa = 9.82GVAWSYY218 pKa = 9.08RR219 pKa = 11.84TVRR222 pKa = 11.84QVLGNVTYY230 pKa = 9.8TGRR233 pKa = 11.84IIFKK237 pKa = 9.61GQVYY241 pKa = 10.45DD242 pKa = 4.04GLHH245 pKa = 5.7TGIISKK251 pKa = 10.64ADD253 pKa = 3.07WDD255 pKa = 3.9EE256 pKa = 3.97VQRR259 pKa = 11.84VLKK262 pKa = 10.31IRR264 pKa = 11.84QLDD267 pKa = 3.57QARR270 pKa = 11.84KK271 pKa = 9.41SNNPRR276 pKa = 11.84PFQARR281 pKa = 11.84YY282 pKa = 8.11MLSGLLKK289 pKa = 10.28CHH291 pKa = 6.0YY292 pKa = 10.11CGATLAIAKK301 pKa = 9.33SHH303 pKa = 5.75TKK305 pKa = 10.38NGPLWRR311 pKa = 11.84YY312 pKa = 7.74VCPTRR317 pKa = 11.84HH318 pKa = 5.33ANKK321 pKa = 9.54YY322 pKa = 8.65HH323 pKa = 6.75RR324 pKa = 11.84NNAARR329 pKa = 11.84YY330 pKa = 7.68RR331 pKa = 11.84IPPVNCEE338 pKa = 3.59LTYY341 pKa = 10.38KK342 pKa = 10.9YY343 pKa = 10.72KK344 pKa = 10.67DD345 pKa = 3.74DD346 pKa = 4.45LEE348 pKa = 4.32EE349 pKa = 4.17AVLRR353 pKa = 11.84EE354 pKa = 4.15VKK356 pKa = 10.26KK357 pKa = 10.98VALDD361 pKa = 3.47PDD363 pKa = 4.08AVVASQDD370 pKa = 3.57DD371 pKa = 4.27GQPKK375 pKa = 9.41IDD377 pKa = 3.62KK378 pKa = 10.3QAIKK382 pKa = 10.6AQLAKK387 pKa = 10.15IKK389 pKa = 10.12RR390 pKa = 11.84QQAKK394 pKa = 10.31LVDD397 pKa = 5.09LYY399 pKa = 11.6LLSDD403 pKa = 4.25DD404 pKa = 5.8LMLTSFTIGPKK415 pKa = 9.1SLKK418 pKa = 10.12SKK420 pKa = 10.6LMLSRR425 pKa = 11.84HH426 pKa = 5.8NLSMMPKK433 pKa = 9.89RR434 pKa = 11.84PTPLKK439 pKa = 9.78RR440 pKa = 11.84RR441 pKa = 11.84RR442 pKa = 11.84PMLHH446 pKa = 6.67RR447 pKa = 11.84LISS450 pKa = 3.52
MM1 pKa = 7.96LEE3 pKa = 3.84VCMKK7 pKa = 8.73IAVYY11 pKa = 10.6VRR13 pKa = 11.84VSTLEE18 pKa = 3.83QAEE21 pKa = 4.25SGYY24 pKa = 10.92SIGEE28 pKa = 3.89QTEE31 pKa = 4.12KK32 pKa = 10.08LTAYY36 pKa = 10.28CRR38 pKa = 11.84VKK40 pKa = 10.25DD41 pKa = 3.54WQVAKK46 pKa = 10.39IYY48 pKa = 9.33TDD50 pKa = 3.52PGFSGSSLDD59 pKa = 3.75RR60 pKa = 11.84PAMQQLIGDD69 pKa = 4.39CKK71 pKa = 10.83LGVFDD76 pKa = 5.52AVLVYY81 pKa = 10.81KK82 pKa = 10.59LDD84 pKa = 3.8RR85 pKa = 11.84LSRR88 pKa = 11.84SQKK91 pKa = 8.91DD92 pKa = 3.35TLYY95 pKa = 11.05LIEE98 pKa = 5.4DD99 pKa = 3.6VFNAYY104 pKa = 10.12NIHH107 pKa = 6.31FMSLSEE113 pKa = 4.46NFDD116 pKa = 3.17TSTPFGKK123 pKa = 10.57AMIGLLSVFAQLEE136 pKa = 4.12RR137 pKa = 11.84EE138 pKa = 4.22QIKK141 pKa = 10.5EE142 pKa = 3.7RR143 pKa = 11.84MQMGKK148 pKa = 10.18LGRR151 pKa = 11.84VKK153 pKa = 10.42AGKK156 pKa = 9.81ISAWSNVPFGYY167 pKa = 9.3TKK169 pKa = 10.82VKK171 pKa = 10.57DD172 pKa = 4.02SYY174 pKa = 11.86DD175 pKa = 3.79VDD177 pKa = 3.89PLRR180 pKa = 11.84ADD182 pKa = 3.3IVRR185 pKa = 11.84RR186 pKa = 11.84IYY188 pKa = 11.28ADD190 pKa = 3.39YY191 pKa = 10.85LAGKK195 pKa = 9.92SITKK199 pKa = 9.68IMKK202 pKa = 8.03EE203 pKa = 4.02LNRR206 pKa = 11.84EE207 pKa = 3.74GHH209 pKa = 5.88IGKK212 pKa = 9.82GVAWSYY218 pKa = 9.08RR219 pKa = 11.84TVRR222 pKa = 11.84QVLGNVTYY230 pKa = 9.8TGRR233 pKa = 11.84IIFKK237 pKa = 9.61GQVYY241 pKa = 10.45DD242 pKa = 4.04GLHH245 pKa = 5.7TGIISKK251 pKa = 10.64ADD253 pKa = 3.07WDD255 pKa = 3.9EE256 pKa = 3.97VQRR259 pKa = 11.84VLKK262 pKa = 10.31IRR264 pKa = 11.84QLDD267 pKa = 3.57QARR270 pKa = 11.84KK271 pKa = 9.41SNNPRR276 pKa = 11.84PFQARR281 pKa = 11.84YY282 pKa = 8.11MLSGLLKK289 pKa = 10.28CHH291 pKa = 6.0YY292 pKa = 10.11CGATLAIAKK301 pKa = 9.33SHH303 pKa = 5.75TKK305 pKa = 10.38NGPLWRR311 pKa = 11.84YY312 pKa = 7.74VCPTRR317 pKa = 11.84HH318 pKa = 5.33ANKK321 pKa = 9.54YY322 pKa = 8.65HH323 pKa = 6.75RR324 pKa = 11.84NNAARR329 pKa = 11.84YY330 pKa = 7.68RR331 pKa = 11.84IPPVNCEE338 pKa = 3.59LTYY341 pKa = 10.38KK342 pKa = 10.9YY343 pKa = 10.72KK344 pKa = 10.67DD345 pKa = 3.74DD346 pKa = 4.45LEE348 pKa = 4.32EE349 pKa = 4.17AVLRR353 pKa = 11.84EE354 pKa = 4.15VKK356 pKa = 10.26KK357 pKa = 10.98VALDD361 pKa = 3.47PDD363 pKa = 4.08AVVASQDD370 pKa = 3.57DD371 pKa = 4.27GQPKK375 pKa = 9.41IDD377 pKa = 3.62KK378 pKa = 10.3QAIKK382 pKa = 10.6AQLAKK387 pKa = 10.15IKK389 pKa = 10.12RR390 pKa = 11.84QQAKK394 pKa = 10.31LVDD397 pKa = 5.09LYY399 pKa = 11.6LLSDD403 pKa = 4.25DD404 pKa = 5.8LMLTSFTIGPKK415 pKa = 9.1SLKK418 pKa = 10.12SKK420 pKa = 10.6LMLSRR425 pKa = 11.84HH426 pKa = 5.8NLSMMPKK433 pKa = 9.89RR434 pKa = 11.84PTPLKK439 pKa = 9.78RR440 pKa = 11.84RR441 pKa = 11.84RR442 pKa = 11.84PMLHH446 pKa = 6.67RR447 pKa = 11.84LISS450 pKa = 3.52
Molecular weight: 51.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10059 |
36 |
966 |
218.7 |
24.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.705 ± 0.453 | 0.557 ± 0.098 |
7.188 ± 0.374 | 5.1 ± 0.446 |
3.539 ± 0.194 | 7.287 ± 0.416 |
1.74 ± 0.163 | 6.541 ± 0.245 |
7.058 ± 0.433 | 7.864 ± 0.299 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.535 ± 0.127 | 5.766 ± 0.224 |
3.519 ± 0.192 | 4.265 ± 0.271 |
4.016 ± 0.3 | 6.601 ± 0.387 |
6.83 ± 0.442 | 6.263 ± 0.242 |
1.66 ± 0.152 | 3.967 ± 0.35 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
