
Pomona leaf-nosed bat associated polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; unclassified Polyomaviridae
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
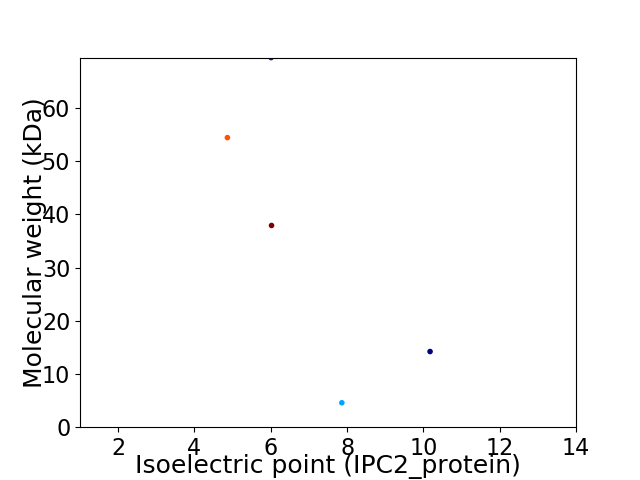
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1R3SYE4|A0A1R3SYE4_9POLY Late orf 1 OS=Pomona leaf-nosed bat associated polyomavirus OX=1885565 GN=LO1 PE=4 SV=1
MM1 pKa = 7.63GIVASIVAAVTGVAEE16 pKa = 4.29AVADD20 pKa = 4.0AAATVAGAAGSAAEE34 pKa = 3.91AAATSFEE41 pKa = 4.42LTSSVGEE48 pKa = 4.07AAGDD52 pKa = 3.85TVPLLDD58 pKa = 4.45GEE60 pKa = 4.54EE61 pKa = 4.04EE62 pKa = 4.68TVFDD66 pKa = 4.48SSWAEE71 pKa = 3.39GGYY74 pKa = 10.65ADD76 pKa = 4.74EE77 pKa = 5.39GAVNDD82 pKa = 3.65PWNWEE87 pKa = 4.01MEE89 pKa = 4.34PEE91 pKa = 4.15DD92 pKa = 4.32LATGSGSRR100 pKa = 11.84LSRR103 pKa = 11.84SLATGLAAGAGVAAAAGGLAWAIGSAAASHH133 pKa = 6.33SEE135 pKa = 3.75AGQALLEE142 pKa = 4.21SSQVIQRR149 pKa = 11.84TLEE152 pKa = 4.02EE153 pKa = 4.32EE154 pKa = 4.77GEE156 pKa = 4.34QPSVPDD162 pKa = 3.12IVMGFMTPEE171 pKa = 3.78EE172 pKa = 4.24YY173 pKa = 10.39EE174 pKa = 3.97RR175 pKa = 11.84ALEE178 pKa = 4.05EE179 pKa = 3.59LHH181 pKa = 6.54RR182 pKa = 11.84RR183 pKa = 11.84QVEE186 pKa = 4.43GGFSPDD192 pKa = 3.1SLRR195 pKa = 11.84EE196 pKa = 3.98AEE198 pKa = 4.12NALVYY203 pKa = 10.17IAPGSGIPYY212 pKa = 9.61RR213 pKa = 11.84VPDD216 pKa = 3.58QSLLMDD222 pKa = 4.41MAQSEE227 pKa = 4.65QPAPWFLPGAAGQPMTRR244 pKa = 11.84GMLRR248 pKa = 11.84RR249 pKa = 11.84AAEE252 pKa = 3.98AARR255 pKa = 11.84VIAEE259 pKa = 4.04GAEE262 pKa = 3.85RR263 pKa = 11.84SQVFRR268 pKa = 11.84LSADD272 pKa = 2.92ARR274 pKa = 11.84RR275 pKa = 11.84EE276 pKa = 3.93LARR279 pKa = 11.84SGQALADD286 pKa = 4.22LYY288 pKa = 11.18RR289 pKa = 11.84RR290 pKa = 11.84GLRR293 pKa = 11.84PSDD296 pKa = 3.39LLYY299 pKa = 10.76RR300 pKa = 11.84QVLRR304 pKa = 11.84GNRR307 pKa = 11.84QVQEE311 pKa = 4.41DD312 pKa = 3.41IARR315 pKa = 11.84RR316 pKa = 11.84VAAEE320 pKa = 3.78IAGQALDD327 pKa = 3.53IGIEE331 pKa = 3.88QGQQIVGNLIGSGVIATSLAAALTGAGHH359 pKa = 7.51LLIDD363 pKa = 4.2KK364 pKa = 10.1YY365 pKa = 10.96FGPSLPAGAVQIEE378 pKa = 4.17NDD380 pKa = 4.38AFDD383 pKa = 3.49QAPNIVRR390 pKa = 11.84ADD392 pKa = 3.19GGLWRR397 pKa = 11.84KK398 pKa = 9.4GRR400 pKa = 11.84ASYY403 pKa = 10.51LVEE406 pKa = 3.85EE407 pKa = 4.71EE408 pKa = 4.52GRR410 pKa = 11.84LGTVDD415 pKa = 3.73LSYY418 pKa = 10.96YY419 pKa = 10.85APEE422 pKa = 4.01TTNRR426 pKa = 11.84GLPSTEE432 pKa = 3.8PWFHH436 pKa = 6.72FPPRR440 pKa = 11.84SPTQLQNLEE449 pKa = 4.38MYY451 pKa = 10.63LKK453 pKa = 10.75NNWGGEE459 pKa = 3.92NTYY462 pKa = 9.75YY463 pKa = 10.66QPTGEE468 pKa = 4.26GTFLIGDD475 pKa = 4.55LVLQEE480 pKa = 3.69AQYY483 pKa = 10.68RR484 pKa = 11.84RR485 pKa = 11.84KK486 pKa = 9.65NARR489 pKa = 11.84AKK491 pKa = 10.21RR492 pKa = 11.84RR493 pKa = 11.84PRR495 pKa = 11.84DD496 pKa = 3.52TGPGTRR502 pKa = 11.84TKK504 pKa = 9.59RR505 pKa = 11.84TRR507 pKa = 11.84RR508 pKa = 11.84ARR510 pKa = 11.84NN511 pKa = 3.05
MM1 pKa = 7.63GIVASIVAAVTGVAEE16 pKa = 4.29AVADD20 pKa = 4.0AAATVAGAAGSAAEE34 pKa = 3.91AAATSFEE41 pKa = 4.42LTSSVGEE48 pKa = 4.07AAGDD52 pKa = 3.85TVPLLDD58 pKa = 4.45GEE60 pKa = 4.54EE61 pKa = 4.04EE62 pKa = 4.68TVFDD66 pKa = 4.48SSWAEE71 pKa = 3.39GGYY74 pKa = 10.65ADD76 pKa = 4.74EE77 pKa = 5.39GAVNDD82 pKa = 3.65PWNWEE87 pKa = 4.01MEE89 pKa = 4.34PEE91 pKa = 4.15DD92 pKa = 4.32LATGSGSRR100 pKa = 11.84LSRR103 pKa = 11.84SLATGLAAGAGVAAAAGGLAWAIGSAAASHH133 pKa = 6.33SEE135 pKa = 3.75AGQALLEE142 pKa = 4.21SSQVIQRR149 pKa = 11.84TLEE152 pKa = 4.02EE153 pKa = 4.32EE154 pKa = 4.77GEE156 pKa = 4.34QPSVPDD162 pKa = 3.12IVMGFMTPEE171 pKa = 3.78EE172 pKa = 4.24YY173 pKa = 10.39EE174 pKa = 3.97RR175 pKa = 11.84ALEE178 pKa = 4.05EE179 pKa = 3.59LHH181 pKa = 6.54RR182 pKa = 11.84RR183 pKa = 11.84QVEE186 pKa = 4.43GGFSPDD192 pKa = 3.1SLRR195 pKa = 11.84EE196 pKa = 3.98AEE198 pKa = 4.12NALVYY203 pKa = 10.17IAPGSGIPYY212 pKa = 9.61RR213 pKa = 11.84VPDD216 pKa = 3.58QSLLMDD222 pKa = 4.41MAQSEE227 pKa = 4.65QPAPWFLPGAAGQPMTRR244 pKa = 11.84GMLRR248 pKa = 11.84RR249 pKa = 11.84AAEE252 pKa = 3.98AARR255 pKa = 11.84VIAEE259 pKa = 4.04GAEE262 pKa = 3.85RR263 pKa = 11.84SQVFRR268 pKa = 11.84LSADD272 pKa = 2.92ARR274 pKa = 11.84RR275 pKa = 11.84EE276 pKa = 3.93LARR279 pKa = 11.84SGQALADD286 pKa = 4.22LYY288 pKa = 11.18RR289 pKa = 11.84RR290 pKa = 11.84GLRR293 pKa = 11.84PSDD296 pKa = 3.39LLYY299 pKa = 10.76RR300 pKa = 11.84QVLRR304 pKa = 11.84GNRR307 pKa = 11.84QVQEE311 pKa = 4.41DD312 pKa = 3.41IARR315 pKa = 11.84RR316 pKa = 11.84VAAEE320 pKa = 3.78IAGQALDD327 pKa = 3.53IGIEE331 pKa = 3.88QGQQIVGNLIGSGVIATSLAAALTGAGHH359 pKa = 7.51LLIDD363 pKa = 4.2KK364 pKa = 10.1YY365 pKa = 10.96FGPSLPAGAVQIEE378 pKa = 4.17NDD380 pKa = 4.38AFDD383 pKa = 3.49QAPNIVRR390 pKa = 11.84ADD392 pKa = 3.19GGLWRR397 pKa = 11.84KK398 pKa = 9.4GRR400 pKa = 11.84ASYY403 pKa = 10.51LVEE406 pKa = 3.85EE407 pKa = 4.71EE408 pKa = 4.52GRR410 pKa = 11.84LGTVDD415 pKa = 3.73LSYY418 pKa = 10.96YY419 pKa = 10.85APEE422 pKa = 4.01TTNRR426 pKa = 11.84GLPSTEE432 pKa = 3.8PWFHH436 pKa = 6.72FPPRR440 pKa = 11.84SPTQLQNLEE449 pKa = 4.38MYY451 pKa = 10.63LKK453 pKa = 10.75NNWGGEE459 pKa = 3.92NTYY462 pKa = 9.75YY463 pKa = 10.66QPTGEE468 pKa = 4.26GTFLIGDD475 pKa = 4.55LVLQEE480 pKa = 3.69AQYY483 pKa = 10.68RR484 pKa = 11.84RR485 pKa = 11.84KK486 pKa = 9.65NARR489 pKa = 11.84AKK491 pKa = 10.21RR492 pKa = 11.84RR493 pKa = 11.84PRR495 pKa = 11.84DD496 pKa = 3.52TGPGTRR502 pKa = 11.84TKK504 pKa = 9.59RR505 pKa = 11.84TRR507 pKa = 11.84RR508 pKa = 11.84ARR510 pKa = 11.84NN511 pKa = 3.05
Molecular weight: 54.42 kDa
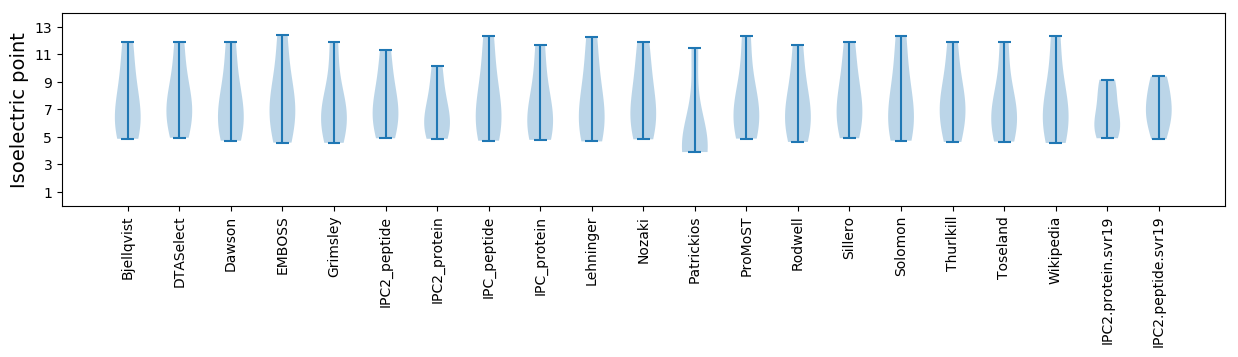
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1R3T5J8|A0A1R3T5J8_9POLY Late orf 2 OS=Pomona leaf-nosed bat associated polyomavirus OX=1885565 GN=LO2 PE=4 SV=1
MM1 pKa = 7.78LLLQLLGLRR10 pKa = 11.84GQLPRR15 pKa = 11.84LLLPALNSLVQLEE28 pKa = 4.58KK29 pKa = 11.22LLGTLFRR36 pKa = 11.84SLMEE40 pKa = 4.02KK41 pKa = 10.1RR42 pKa = 11.84KK43 pKa = 9.91QSSTPLGPRR52 pKa = 11.84GAMLMRR58 pKa = 11.84GLLMTRR64 pKa = 11.84GTGKK68 pKa = 9.78WNLRR72 pKa = 11.84TWLLVVAADD81 pKa = 5.04CPDD84 pKa = 3.77LLLPAWQLGQEE95 pKa = 3.98WQLPRR100 pKa = 11.84GAWPGQLVRR109 pKa = 11.84LLLLILRR116 pKa = 11.84LVKK119 pKa = 10.4PSLRR123 pKa = 11.84ALKK126 pKa = 10.49
MM1 pKa = 7.78LLLQLLGLRR10 pKa = 11.84GQLPRR15 pKa = 11.84LLLPALNSLVQLEE28 pKa = 4.58KK29 pKa = 11.22LLGTLFRR36 pKa = 11.84SLMEE40 pKa = 4.02KK41 pKa = 10.1RR42 pKa = 11.84KK43 pKa = 9.91QSSTPLGPRR52 pKa = 11.84GAMLMRR58 pKa = 11.84GLLMTRR64 pKa = 11.84GTGKK68 pKa = 9.78WNLRR72 pKa = 11.84TWLLVVAADD81 pKa = 5.04CPDD84 pKa = 3.77LLLPAWQLGQEE95 pKa = 3.98WQLPRR100 pKa = 11.84GAWPGQLVRR109 pKa = 11.84LLLLILRR116 pKa = 11.84LVKK119 pKa = 10.4PSLRR123 pKa = 11.84ALKK126 pKa = 10.49
Molecular weight: 14.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1645 |
44 |
616 |
329.0 |
36.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.119 ± 2.208 | 1.337 ± 0.511 |
4.681 ± 0.798 | 6.261 ± 1.056 |
4.134 ± 1.108 | 8.693 ± 1.164 |
1.824 ± 0.688 | 3.04 ± 0.303 |
4.012 ± 1.096 | 11.003 ± 1.878 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.614 ± 0.327 | 3.647 ± 0.799 |
6.383 ± 0.739 | 4.377 ± 0.426 |
6.261 ± 1.119 | 7.234 ± 0.578 |
5.532 ± 0.975 | 5.775 ± 0.587 |
1.641 ± 0.353 | 2.432 ± 0.285 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
