
Pythium nunn virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Gammapartitivirus; unclassified Gammapartitivirus
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
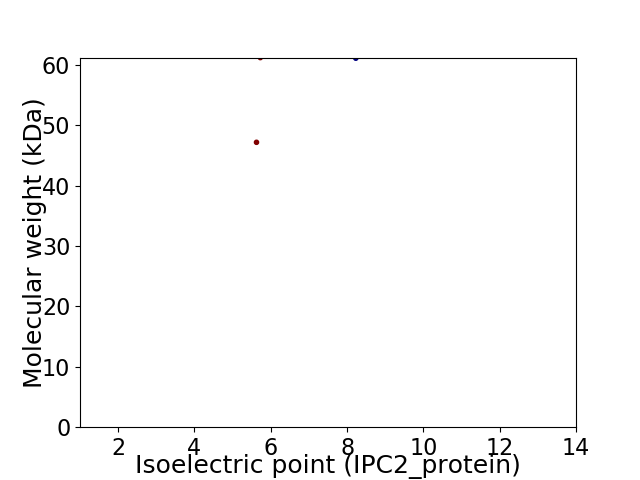
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5ZAM9|A0A2Z5ZAM9_9VIRU Putative capsid protein OS=Pythium nunn virus 1 OX=2083275 GN=CP PE=4 SV=1
MM1 pKa = 7.68NYY3 pKa = 9.83NARR6 pKa = 11.84DD7 pKa = 3.43VGEE10 pKa = 4.28AVSNGQPPRR19 pKa = 11.84RR20 pKa = 11.84AGDD23 pKa = 3.64NPNKK27 pKa = 10.42AFAQRR32 pKa = 11.84RR33 pKa = 11.84QGNRR37 pKa = 11.84PKK39 pKa = 10.83VKK41 pKa = 9.1NTPSGPKK48 pKa = 7.61TTTEE52 pKa = 3.57AFRR55 pKa = 11.84WIEE58 pKa = 3.98PLPEE62 pKa = 4.18VTPMPAVLEE71 pKa = 4.65TITEE75 pKa = 4.45SIPAGSIEE83 pKa = 5.22LNFEE87 pKa = 4.26LPSTVASPFTQSVEE101 pKa = 4.28SVLMRR106 pKa = 11.84TDD108 pKa = 3.84LPSASIYY115 pKa = 7.73QARR118 pKa = 11.84DD119 pKa = 3.06KK120 pKa = 11.56LEE122 pKa = 3.97ALSYY126 pKa = 11.02YY127 pKa = 9.79KK128 pKa = 10.4ACRR131 pKa = 11.84QLYY134 pKa = 8.14STLSDD139 pKa = 3.59PQKK142 pKa = 11.05AALQPLKK149 pKa = 10.65AIYY152 pKa = 9.49YY153 pKa = 8.49DD154 pKa = 3.63KK155 pKa = 11.42SHH157 pKa = 6.98IPNHH161 pKa = 5.51MSAALSMIGNLEE173 pKa = 4.13SKK175 pKa = 9.92MGLIEE180 pKa = 4.3VKK182 pKa = 10.14HH183 pKa = 6.19APVLFKK189 pKa = 10.55RR190 pKa = 11.84WLIAGLCIDD199 pKa = 4.9PDD201 pKa = 3.96TTYY204 pKa = 10.42TAATVAPNSLVFKK217 pKa = 10.44DD218 pKa = 4.29VYY220 pKa = 11.01SKK222 pKa = 11.7ALIDD226 pKa = 3.61EE227 pKa = 4.63KK228 pKa = 11.39ANEE231 pKa = 4.06RR232 pKa = 11.84LKK234 pKa = 10.62EE235 pKa = 3.85IHH237 pKa = 6.57NEE239 pKa = 3.82TFKK242 pKa = 11.04IKK244 pKa = 10.7VGTTDD249 pKa = 3.37IEE251 pKa = 4.48VSAPIPPPTDD261 pKa = 2.77AGYY264 pKa = 11.4NSIPDD269 pKa = 4.53DD270 pKa = 3.93YY271 pKa = 11.23PHH273 pKa = 7.16ADD275 pKa = 3.23EE276 pKa = 5.08MRR278 pKa = 11.84DD279 pKa = 3.45LVSIIQMATRR289 pKa = 11.84DD290 pKa = 4.0YY291 pKa = 11.28VRR293 pKa = 11.84SQPVPHH299 pKa = 6.79GRR301 pKa = 11.84DD302 pKa = 3.03LDD304 pKa = 3.99TALSHH309 pKa = 7.18LGLQVAPAGYY319 pKa = 9.98SDD321 pKa = 3.52QDD323 pKa = 3.31LRR325 pKa = 11.84TAFEE329 pKa = 4.5DD330 pKa = 4.74AIEE333 pKa = 4.55TYY335 pKa = 9.22LTTEE339 pKa = 4.68IIHH342 pKa = 6.01IQTVFHH348 pKa = 6.66LGEE351 pKa = 4.42PPATEE356 pKa = 4.12RR357 pKa = 11.84GFASQLVASDD367 pKa = 4.07DD368 pKa = 3.45VTARR372 pKa = 11.84FGLPLSDD379 pKa = 3.98ADD381 pKa = 3.76KK382 pKa = 11.48ALGFMFNPRR391 pKa = 11.84HH392 pKa = 5.53SFEE395 pKa = 4.18FNPRR399 pKa = 11.84FVAYY403 pKa = 9.87SRR405 pKa = 11.84RR406 pKa = 11.84AKK408 pKa = 9.56RR409 pKa = 11.84TTASQFASNDD419 pKa = 3.08LRR421 pKa = 11.84HH422 pKa = 6.6IGTDD426 pKa = 3.0
MM1 pKa = 7.68NYY3 pKa = 9.83NARR6 pKa = 11.84DD7 pKa = 3.43VGEE10 pKa = 4.28AVSNGQPPRR19 pKa = 11.84RR20 pKa = 11.84AGDD23 pKa = 3.64NPNKK27 pKa = 10.42AFAQRR32 pKa = 11.84RR33 pKa = 11.84QGNRR37 pKa = 11.84PKK39 pKa = 10.83VKK41 pKa = 9.1NTPSGPKK48 pKa = 7.61TTTEE52 pKa = 3.57AFRR55 pKa = 11.84WIEE58 pKa = 3.98PLPEE62 pKa = 4.18VTPMPAVLEE71 pKa = 4.65TITEE75 pKa = 4.45SIPAGSIEE83 pKa = 5.22LNFEE87 pKa = 4.26LPSTVASPFTQSVEE101 pKa = 4.28SVLMRR106 pKa = 11.84TDD108 pKa = 3.84LPSASIYY115 pKa = 7.73QARR118 pKa = 11.84DD119 pKa = 3.06KK120 pKa = 11.56LEE122 pKa = 3.97ALSYY126 pKa = 11.02YY127 pKa = 9.79KK128 pKa = 10.4ACRR131 pKa = 11.84QLYY134 pKa = 8.14STLSDD139 pKa = 3.59PQKK142 pKa = 11.05AALQPLKK149 pKa = 10.65AIYY152 pKa = 9.49YY153 pKa = 8.49DD154 pKa = 3.63KK155 pKa = 11.42SHH157 pKa = 6.98IPNHH161 pKa = 5.51MSAALSMIGNLEE173 pKa = 4.13SKK175 pKa = 9.92MGLIEE180 pKa = 4.3VKK182 pKa = 10.14HH183 pKa = 6.19APVLFKK189 pKa = 10.55RR190 pKa = 11.84WLIAGLCIDD199 pKa = 4.9PDD201 pKa = 3.96TTYY204 pKa = 10.42TAATVAPNSLVFKK217 pKa = 10.44DD218 pKa = 4.29VYY220 pKa = 11.01SKK222 pKa = 11.7ALIDD226 pKa = 3.61EE227 pKa = 4.63KK228 pKa = 11.39ANEE231 pKa = 4.06RR232 pKa = 11.84LKK234 pKa = 10.62EE235 pKa = 3.85IHH237 pKa = 6.57NEE239 pKa = 3.82TFKK242 pKa = 11.04IKK244 pKa = 10.7VGTTDD249 pKa = 3.37IEE251 pKa = 4.48VSAPIPPPTDD261 pKa = 2.77AGYY264 pKa = 11.4NSIPDD269 pKa = 4.53DD270 pKa = 3.93YY271 pKa = 11.23PHH273 pKa = 7.16ADD275 pKa = 3.23EE276 pKa = 5.08MRR278 pKa = 11.84DD279 pKa = 3.45LVSIIQMATRR289 pKa = 11.84DD290 pKa = 4.0YY291 pKa = 11.28VRR293 pKa = 11.84SQPVPHH299 pKa = 6.79GRR301 pKa = 11.84DD302 pKa = 3.03LDD304 pKa = 3.99TALSHH309 pKa = 7.18LGLQVAPAGYY319 pKa = 9.98SDD321 pKa = 3.52QDD323 pKa = 3.31LRR325 pKa = 11.84TAFEE329 pKa = 4.5DD330 pKa = 4.74AIEE333 pKa = 4.55TYY335 pKa = 9.22LTTEE339 pKa = 4.68IIHH342 pKa = 6.01IQTVFHH348 pKa = 6.66LGEE351 pKa = 4.42PPATEE356 pKa = 4.12RR357 pKa = 11.84GFASQLVASDD367 pKa = 4.07DD368 pKa = 3.45VTARR372 pKa = 11.84FGLPLSDD379 pKa = 3.98ADD381 pKa = 3.76KK382 pKa = 11.48ALGFMFNPRR391 pKa = 11.84HH392 pKa = 5.53SFEE395 pKa = 4.18FNPRR399 pKa = 11.84FVAYY403 pKa = 9.87SRR405 pKa = 11.84RR406 pKa = 11.84AKK408 pKa = 9.56RR409 pKa = 11.84TTASQFASNDD419 pKa = 3.08LRR421 pKa = 11.84HH422 pKa = 6.6IGTDD426 pKa = 3.0
Molecular weight: 47.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5ZAM9|A0A2Z5ZAM9_9VIRU Putative capsid protein OS=Pythium nunn virus 1 OX=2083275 GN=CP PE=4 SV=1
MM1 pKa = 6.44TTKK4 pKa = 9.37KK5 pKa = 8.77TRR7 pKa = 11.84AEE9 pKa = 3.85VQEE12 pKa = 3.98FLKK15 pKa = 10.36TIAGKK20 pKa = 9.51PGLRR24 pKa = 11.84VVPDD28 pKa = 4.26PNAYY32 pKa = 9.02PYY34 pKa = 10.61NVEE37 pKa = 4.08EE38 pKa = 4.35TRR40 pKa = 11.84MNTDD44 pKa = 3.02PFVRR48 pKa = 11.84KK49 pKa = 9.74SMKK52 pKa = 8.84LWSRR56 pKa = 11.84EE57 pKa = 3.69TYY59 pKa = 8.36EE60 pKa = 3.99AQRR63 pKa = 11.84GFTKK67 pKa = 10.58RR68 pKa = 11.84ATLSNSLYY76 pKa = 9.97TLQKK80 pKa = 8.74YY81 pKa = 9.89ASPQRR86 pKa = 11.84LKK88 pKa = 11.03SQISHH93 pKa = 5.97EE94 pKa = 4.25FEE96 pKa = 4.22SCIEE100 pKa = 4.01SAIAYY105 pKa = 7.45ARR107 pKa = 11.84HH108 pKa = 5.34VFIPQDD114 pKa = 3.05KK115 pKa = 10.1LLRR118 pKa = 11.84YY119 pKa = 9.4SLAHH123 pKa = 6.02GTDD126 pKa = 3.0NMNLDD131 pKa = 3.56SAAGFSYY138 pKa = 10.75PGMKK142 pKa = 9.65KK143 pKa = 10.47AEE145 pKa = 4.33VVEE148 pKa = 4.17EE149 pKa = 4.63AYY151 pKa = 10.44DD152 pKa = 3.5VAAYY156 pKa = 8.71LQHH159 pKa = 5.05MTQRR163 pKa = 11.84GKK165 pKa = 10.35SVYY168 pKa = 10.01IPPCKK173 pKa = 9.79VALRR177 pKa = 11.84GHH179 pKa = 7.15LSSQEE184 pKa = 4.07EE185 pKa = 4.33QKK187 pKa = 10.86SRR189 pKa = 11.84PVWVYY194 pKa = 9.26PVEE197 pKa = 4.22VTILEE202 pKa = 4.51SKK204 pKa = 9.14WAIPFYY210 pKa = 10.78EE211 pKa = 4.04HH212 pKa = 7.6LEE214 pKa = 4.29RR215 pKa = 11.84NVEE218 pKa = 4.34TVHH221 pKa = 6.79FGANAMPKK229 pKa = 10.37LSQLLMGGLADD240 pKa = 3.61HH241 pKa = 6.9SEE243 pKa = 4.08AAEE246 pKa = 4.08ITLDD250 pKa = 3.13WSNFDD255 pKa = 3.58SSIPNWLINQAFDD268 pKa = 4.85IIWEE272 pKa = 4.32SFDD275 pKa = 3.26NQYY278 pKa = 11.41AYY280 pKa = 10.66HH281 pKa = 6.83EE282 pKa = 4.89GEE284 pKa = 3.76AVYY287 pKa = 10.28GGEE290 pKa = 3.93VMEE293 pKa = 4.95RR294 pKa = 11.84KK295 pKa = 9.8NINLFRR301 pKa = 11.84WIKK304 pKa = 10.37EE305 pKa = 3.61YY306 pKa = 10.75FINTKK311 pKa = 10.1IMLPSGDD318 pKa = 3.69VVRR321 pKa = 11.84KK322 pKa = 6.49THH324 pKa = 7.05GIPSGSFFTQAVGSIVNYY342 pKa = 10.35IMVKK346 pKa = 10.57ALDD349 pKa = 5.03LYY351 pKa = 10.26FSWGARR357 pKa = 11.84RR358 pKa = 11.84IRR360 pKa = 11.84ILGDD364 pKa = 3.23DD365 pKa = 3.98SSFLIPLFSEE375 pKa = 4.81KK376 pKa = 10.24KK377 pKa = 10.03CEE379 pKa = 3.98AKK381 pKa = 10.5AIQNAAWKK389 pKa = 10.49AFGVTLKK396 pKa = 10.71LPKK399 pKa = 10.21LRR401 pKa = 11.84ISTTQKK407 pKa = 8.99QRR409 pKa = 11.84KK410 pKa = 7.56FLGYY414 pKa = 10.19QITGHH419 pKa = 6.59RR420 pKa = 11.84LFRR423 pKa = 11.84DD424 pKa = 3.21DD425 pKa = 6.49DD426 pKa = 3.98EE427 pKa = 4.41FLKK430 pKa = 11.06LVLYY434 pKa = 8.8PEE436 pKa = 4.93RR437 pKa = 11.84DD438 pKa = 3.55VEE440 pKa = 4.31TLEE443 pKa = 4.15QSASRR448 pKa = 11.84VLAYY452 pKa = 11.12YY453 pKa = 9.8MLGGVNDD460 pKa = 3.78EE461 pKa = 4.85VYY463 pKa = 10.98CSFFWDD469 pKa = 3.5YY470 pKa = 11.17LGRR473 pKa = 11.84YY474 pKa = 6.48PAVIGRR480 pKa = 11.84EE481 pKa = 3.97LRR483 pKa = 11.84LTRR486 pKa = 11.84GMKK489 pKa = 10.34RR490 pKa = 11.84MFKK493 pKa = 10.02FVLRR497 pKa = 11.84QEE499 pKa = 4.21VDD501 pKa = 3.39VLRR504 pKa = 11.84VPDD507 pKa = 4.2FTKK510 pKa = 10.71LDD512 pKa = 3.84PTFVPYY518 pKa = 11.24LMTLGDD524 pKa = 4.5RR525 pKa = 11.84PFSSNN530 pKa = 3.07
MM1 pKa = 6.44TTKK4 pKa = 9.37KK5 pKa = 8.77TRR7 pKa = 11.84AEE9 pKa = 3.85VQEE12 pKa = 3.98FLKK15 pKa = 10.36TIAGKK20 pKa = 9.51PGLRR24 pKa = 11.84VVPDD28 pKa = 4.26PNAYY32 pKa = 9.02PYY34 pKa = 10.61NVEE37 pKa = 4.08EE38 pKa = 4.35TRR40 pKa = 11.84MNTDD44 pKa = 3.02PFVRR48 pKa = 11.84KK49 pKa = 9.74SMKK52 pKa = 8.84LWSRR56 pKa = 11.84EE57 pKa = 3.69TYY59 pKa = 8.36EE60 pKa = 3.99AQRR63 pKa = 11.84GFTKK67 pKa = 10.58RR68 pKa = 11.84ATLSNSLYY76 pKa = 9.97TLQKK80 pKa = 8.74YY81 pKa = 9.89ASPQRR86 pKa = 11.84LKK88 pKa = 11.03SQISHH93 pKa = 5.97EE94 pKa = 4.25FEE96 pKa = 4.22SCIEE100 pKa = 4.01SAIAYY105 pKa = 7.45ARR107 pKa = 11.84HH108 pKa = 5.34VFIPQDD114 pKa = 3.05KK115 pKa = 10.1LLRR118 pKa = 11.84YY119 pKa = 9.4SLAHH123 pKa = 6.02GTDD126 pKa = 3.0NMNLDD131 pKa = 3.56SAAGFSYY138 pKa = 10.75PGMKK142 pKa = 9.65KK143 pKa = 10.47AEE145 pKa = 4.33VVEE148 pKa = 4.17EE149 pKa = 4.63AYY151 pKa = 10.44DD152 pKa = 3.5VAAYY156 pKa = 8.71LQHH159 pKa = 5.05MTQRR163 pKa = 11.84GKK165 pKa = 10.35SVYY168 pKa = 10.01IPPCKK173 pKa = 9.79VALRR177 pKa = 11.84GHH179 pKa = 7.15LSSQEE184 pKa = 4.07EE185 pKa = 4.33QKK187 pKa = 10.86SRR189 pKa = 11.84PVWVYY194 pKa = 9.26PVEE197 pKa = 4.22VTILEE202 pKa = 4.51SKK204 pKa = 9.14WAIPFYY210 pKa = 10.78EE211 pKa = 4.04HH212 pKa = 7.6LEE214 pKa = 4.29RR215 pKa = 11.84NVEE218 pKa = 4.34TVHH221 pKa = 6.79FGANAMPKK229 pKa = 10.37LSQLLMGGLADD240 pKa = 3.61HH241 pKa = 6.9SEE243 pKa = 4.08AAEE246 pKa = 4.08ITLDD250 pKa = 3.13WSNFDD255 pKa = 3.58SSIPNWLINQAFDD268 pKa = 4.85IIWEE272 pKa = 4.32SFDD275 pKa = 3.26NQYY278 pKa = 11.41AYY280 pKa = 10.66HH281 pKa = 6.83EE282 pKa = 4.89GEE284 pKa = 3.76AVYY287 pKa = 10.28GGEE290 pKa = 3.93VMEE293 pKa = 4.95RR294 pKa = 11.84KK295 pKa = 9.8NINLFRR301 pKa = 11.84WIKK304 pKa = 10.37EE305 pKa = 3.61YY306 pKa = 10.75FINTKK311 pKa = 10.1IMLPSGDD318 pKa = 3.69VVRR321 pKa = 11.84KK322 pKa = 6.49THH324 pKa = 7.05GIPSGSFFTQAVGSIVNYY342 pKa = 10.35IMVKK346 pKa = 10.57ALDD349 pKa = 5.03LYY351 pKa = 10.26FSWGARR357 pKa = 11.84RR358 pKa = 11.84IRR360 pKa = 11.84ILGDD364 pKa = 3.23DD365 pKa = 3.98SSFLIPLFSEE375 pKa = 4.81KK376 pKa = 10.24KK377 pKa = 10.03CEE379 pKa = 3.98AKK381 pKa = 10.5AIQNAAWKK389 pKa = 10.49AFGVTLKK396 pKa = 10.71LPKK399 pKa = 10.21LRR401 pKa = 11.84ISTTQKK407 pKa = 8.99QRR409 pKa = 11.84KK410 pKa = 7.56FLGYY414 pKa = 10.19QITGHH419 pKa = 6.59RR420 pKa = 11.84LFRR423 pKa = 11.84DD424 pKa = 3.21DD425 pKa = 6.49DD426 pKa = 3.98EE427 pKa = 4.41FLKK430 pKa = 11.06LVLYY434 pKa = 8.8PEE436 pKa = 4.93RR437 pKa = 11.84DD438 pKa = 3.55VEE440 pKa = 4.31TLEE443 pKa = 4.15QSASRR448 pKa = 11.84VLAYY452 pKa = 11.12YY453 pKa = 9.8MLGGVNDD460 pKa = 3.78EE461 pKa = 4.85VYY463 pKa = 10.98CSFFWDD469 pKa = 3.5YY470 pKa = 11.17LGRR473 pKa = 11.84YY474 pKa = 6.48PAVIGRR480 pKa = 11.84EE481 pKa = 3.97LRR483 pKa = 11.84LTRR486 pKa = 11.84GMKK489 pKa = 10.34RR490 pKa = 11.84MFKK493 pKa = 10.02FVLRR497 pKa = 11.84QEE499 pKa = 4.21VDD501 pKa = 3.39VLRR504 pKa = 11.84VPDD507 pKa = 4.2FTKK510 pKa = 10.71LDD512 pKa = 3.84PTFVPYY518 pKa = 11.24LMTLGDD524 pKa = 4.5RR525 pKa = 11.84PFSSNN530 pKa = 3.07
Molecular weight: 61.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
956 |
426 |
530 |
478.0 |
54.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.473 ± 1.267 | 0.628 ± 0.108 |
5.544 ± 0.703 | 6.276 ± 0.439 |
4.812 ± 0.561 | 5.021 ± 0.383 |
2.301 ± 0.192 | 5.335 ± 0.204 |
5.649 ± 0.651 | 8.577 ± 0.407 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.51 ± 0.272 | 3.766 ± 0.154 |
6.172 ± 1.236 | 3.661 ± 0.096 |
6.172 ± 0.207 | 7.218 ± 0.201 |
6.067 ± 0.826 | 6.172 ± 0.527 |
1.255 ± 0.537 | 4.393 ± 0.596 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
