
Haloterrigena turkmenica (strain ATCC 51198 / DSM 5511 / JCM 9101 / NCIMB 13204 / VKM B-1734 / 4k) (Halococcus turkmenicus)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Haloterrigena; Haloterrigena turkmenica
Average proteome isoelectric point is 4.78
Get precalculated fractions of proteins
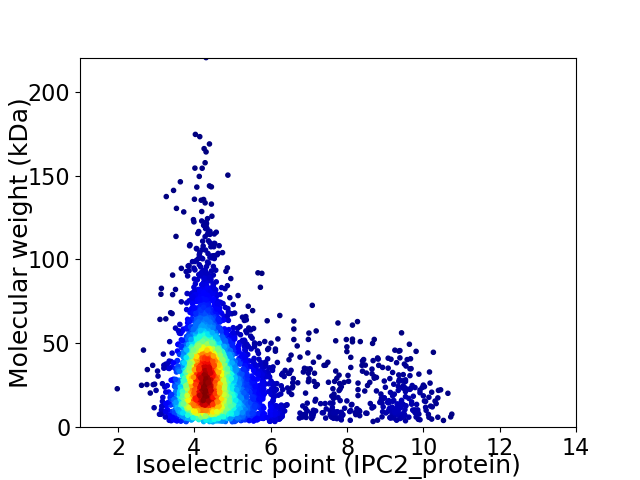
Virtual 2D-PAGE plot for 5113 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D2S1U6|D2S1U6_HALTV Uncharacterized protein OS=Haloterrigena turkmenica (strain ATCC 51198 / DSM 5511 / JCM 9101 / NCIMB 13204 / VKM B-1734 / 4k) OX=543526 GN=Htur_4539 PE=4 SV=1
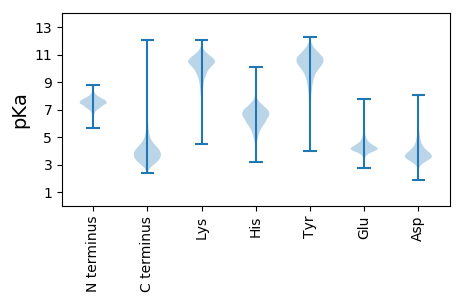
MM1 pKa = 7.02KK2 pKa = 9.23QTRR5 pKa = 11.84RR6 pKa = 11.84TYY8 pKa = 11.14LKK10 pKa = 9.28GTAASALIGIGALSGLSGSAAAEE33 pKa = 4.31SNFDD37 pKa = 4.07LEE39 pKa = 5.47AGFADD44 pKa = 4.89TSWLDD49 pKa = 3.49DD50 pKa = 4.28DD51 pKa = 5.63VDD53 pKa = 3.71VHH55 pKa = 6.73TITEE59 pKa = 4.21PTRR62 pKa = 11.84SAVEE66 pKa = 4.16SAFSASGARR75 pKa = 11.84VVVFEE80 pKa = 4.2TSGTIDD86 pKa = 4.12LGGNDD91 pKa = 3.76LAITEE96 pKa = 4.56DD97 pKa = 4.19YY98 pKa = 10.75CWVAGQTAPSPGITFINGQVRR119 pKa = 11.84ISANNCVVQHH129 pKa = 5.23IRR131 pKa = 11.84SRR133 pKa = 11.84IGPGSDD139 pKa = 4.29GSIQSNDD146 pKa = 3.3AFNTADD152 pKa = 3.41GTQNNVVDD160 pKa = 4.58HH161 pKa = 6.64VSASWGTDD169 pKa = 2.56EE170 pKa = 4.91CLSVGYY176 pKa = 8.02DD177 pKa = 3.52TQDD180 pKa = 3.14TTVTNCLIYY189 pKa = 10.56EE190 pKa = 4.46GLYY193 pKa = 10.7DD194 pKa = 4.06PYY196 pKa = 11.51GNEE199 pKa = 3.63ADD201 pKa = 3.99HH202 pKa = 7.08NYY204 pKa = 10.53GSLIGDD210 pKa = 4.09GASNVTLAGNVWGKK224 pKa = 10.05VRR226 pKa = 11.84GRR228 pKa = 11.84APRR231 pKa = 11.84LKK233 pKa = 10.37SDD235 pKa = 3.51TEE237 pKa = 4.35TVVVNNLLYY246 pKa = 10.88FFDD249 pKa = 4.26EE250 pKa = 4.56SANADD255 pKa = 3.66DD256 pKa = 4.24SAVTSFVGNAAICADD271 pKa = 4.48DD272 pKa = 5.56DD273 pKa = 4.28DD274 pKa = 7.06AILEE278 pKa = 4.33GSPTAYY284 pKa = 10.31HH285 pKa = 7.33ADD287 pKa = 3.71NIAYY291 pKa = 7.97DD292 pKa = 3.84PPMVDD297 pKa = 3.52EE298 pKa = 4.56QPIAEE303 pKa = 4.56PEE305 pKa = 4.5STSSPPLWPSGLSEE319 pKa = 4.29MPSGDD324 pKa = 3.73VEE326 pKa = 4.73SHH328 pKa = 6.05NLTNAGARR336 pKa = 11.84PADD339 pKa = 3.83RR340 pKa = 11.84TQNDD344 pKa = 2.79ARR346 pKa = 11.84IVQEE350 pKa = 3.64IADD353 pKa = 4.53RR354 pKa = 11.84AGLDD358 pKa = 3.96YY359 pKa = 11.19LDD361 pKa = 4.52SPYY364 pKa = 10.89DD365 pKa = 3.56YY366 pKa = 10.53WVGHH370 pKa = 6.24HH371 pKa = 7.35DD372 pKa = 3.85EE373 pKa = 4.38VGGYY377 pKa = 9.56PEE379 pKa = 5.64LPVNTHH385 pKa = 5.34SLEE388 pKa = 4.26VPDD391 pKa = 4.57SGIRR395 pKa = 11.84DD396 pKa = 3.82WLAGWAQAVEE406 pKa = 4.72EE407 pKa = 5.08GSSPPDD413 pKa = 3.63GGSGDD418 pKa = 4.62DD419 pKa = 4.28GSSGPIPTGTYY430 pKa = 9.85EE431 pKa = 3.87IANVNSGQLLEE442 pKa = 4.4VADD445 pKa = 4.56ASTADD450 pKa = 3.76GANVQQWSATDD461 pKa = 3.35HH462 pKa = 6.7ATQQWYY468 pKa = 10.41VEE470 pKa = 4.03DD471 pKa = 3.96TGNGEE476 pKa = 4.36YY477 pKa = 10.78VLQNANSGLLLEE489 pKa = 4.74VADD492 pKa = 5.31GSTEE496 pKa = 4.05DD497 pKa = 3.65GANVQQHH504 pKa = 6.9ADD506 pKa = 3.74TGCDD510 pKa = 3.4CQRR513 pKa = 11.84WSINDD518 pKa = 3.42VGNGEE523 pKa = 4.51YY524 pKa = 10.02ILEE527 pKa = 4.38AVHH530 pKa = 6.19SGKK533 pKa = 10.32VADD536 pKa = 4.19VEE538 pKa = 4.62GASTSDD544 pKa = 3.37GANVLQWPDD553 pKa = 2.89TGGANQRR560 pKa = 11.84WTFDD564 pKa = 3.43SVV566 pKa = 2.95
MM1 pKa = 7.02KK2 pKa = 9.23QTRR5 pKa = 11.84RR6 pKa = 11.84TYY8 pKa = 11.14LKK10 pKa = 9.28GTAASALIGIGALSGLSGSAAAEE33 pKa = 4.31SNFDD37 pKa = 4.07LEE39 pKa = 5.47AGFADD44 pKa = 4.89TSWLDD49 pKa = 3.49DD50 pKa = 4.28DD51 pKa = 5.63VDD53 pKa = 3.71VHH55 pKa = 6.73TITEE59 pKa = 4.21PTRR62 pKa = 11.84SAVEE66 pKa = 4.16SAFSASGARR75 pKa = 11.84VVVFEE80 pKa = 4.2TSGTIDD86 pKa = 4.12LGGNDD91 pKa = 3.76LAITEE96 pKa = 4.56DD97 pKa = 4.19YY98 pKa = 10.75CWVAGQTAPSPGITFINGQVRR119 pKa = 11.84ISANNCVVQHH129 pKa = 5.23IRR131 pKa = 11.84SRR133 pKa = 11.84IGPGSDD139 pKa = 4.29GSIQSNDD146 pKa = 3.3AFNTADD152 pKa = 3.41GTQNNVVDD160 pKa = 4.58HH161 pKa = 6.64VSASWGTDD169 pKa = 2.56EE170 pKa = 4.91CLSVGYY176 pKa = 8.02DD177 pKa = 3.52TQDD180 pKa = 3.14TTVTNCLIYY189 pKa = 10.56EE190 pKa = 4.46GLYY193 pKa = 10.7DD194 pKa = 4.06PYY196 pKa = 11.51GNEE199 pKa = 3.63ADD201 pKa = 3.99HH202 pKa = 7.08NYY204 pKa = 10.53GSLIGDD210 pKa = 4.09GASNVTLAGNVWGKK224 pKa = 10.05VRR226 pKa = 11.84GRR228 pKa = 11.84APRR231 pKa = 11.84LKK233 pKa = 10.37SDD235 pKa = 3.51TEE237 pKa = 4.35TVVVNNLLYY246 pKa = 10.88FFDD249 pKa = 4.26EE250 pKa = 4.56SANADD255 pKa = 3.66DD256 pKa = 4.24SAVTSFVGNAAICADD271 pKa = 4.48DD272 pKa = 5.56DD273 pKa = 4.28DD274 pKa = 7.06AILEE278 pKa = 4.33GSPTAYY284 pKa = 10.31HH285 pKa = 7.33ADD287 pKa = 3.71NIAYY291 pKa = 7.97DD292 pKa = 3.84PPMVDD297 pKa = 3.52EE298 pKa = 4.56QPIAEE303 pKa = 4.56PEE305 pKa = 4.5STSSPPLWPSGLSEE319 pKa = 4.29MPSGDD324 pKa = 3.73VEE326 pKa = 4.73SHH328 pKa = 6.05NLTNAGARR336 pKa = 11.84PADD339 pKa = 3.83RR340 pKa = 11.84TQNDD344 pKa = 2.79ARR346 pKa = 11.84IVQEE350 pKa = 3.64IADD353 pKa = 4.53RR354 pKa = 11.84AGLDD358 pKa = 3.96YY359 pKa = 11.19LDD361 pKa = 4.52SPYY364 pKa = 10.89DD365 pKa = 3.56YY366 pKa = 10.53WVGHH370 pKa = 6.24HH371 pKa = 7.35DD372 pKa = 3.85EE373 pKa = 4.38VGGYY377 pKa = 9.56PEE379 pKa = 5.64LPVNTHH385 pKa = 5.34SLEE388 pKa = 4.26VPDD391 pKa = 4.57SGIRR395 pKa = 11.84DD396 pKa = 3.82WLAGWAQAVEE406 pKa = 4.72EE407 pKa = 5.08GSSPPDD413 pKa = 3.63GGSGDD418 pKa = 4.62DD419 pKa = 4.28GSSGPIPTGTYY430 pKa = 9.85EE431 pKa = 3.87IANVNSGQLLEE442 pKa = 4.4VADD445 pKa = 4.56ASTADD450 pKa = 3.76GANVQQWSATDD461 pKa = 3.35HH462 pKa = 6.7ATQQWYY468 pKa = 10.41VEE470 pKa = 4.03DD471 pKa = 3.96TGNGEE476 pKa = 4.36YY477 pKa = 10.78VLQNANSGLLLEE489 pKa = 4.74VADD492 pKa = 5.31GSTEE496 pKa = 4.05DD497 pKa = 3.65GANVQQHH504 pKa = 6.9ADD506 pKa = 3.74TGCDD510 pKa = 3.4CQRR513 pKa = 11.84WSINDD518 pKa = 3.42VGNGEE523 pKa = 4.51YY524 pKa = 10.02ILEE527 pKa = 4.38AVHH530 pKa = 6.19SGKK533 pKa = 10.32VADD536 pKa = 4.19VEE538 pKa = 4.62GASTSDD544 pKa = 3.37GANVLQWPDD553 pKa = 2.89TGGANQRR560 pKa = 11.84WTFDD564 pKa = 3.43SVV566 pKa = 2.95
Molecular weight: 59.58 kDa
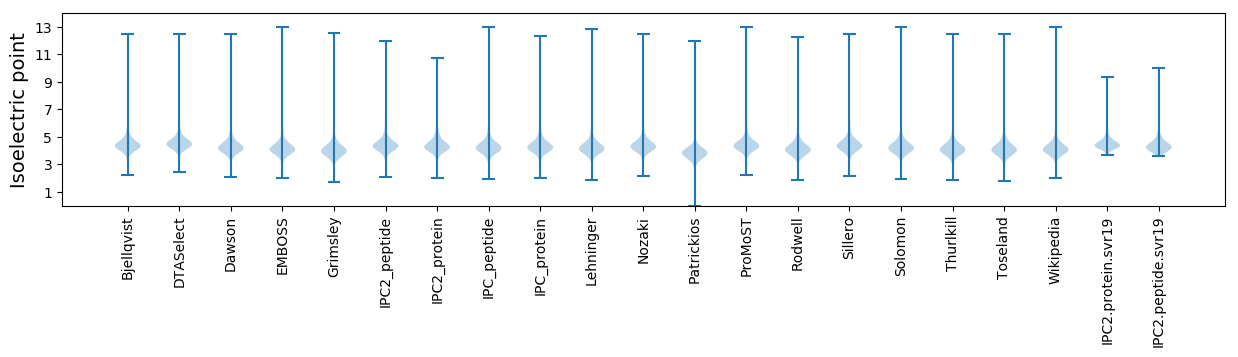
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D2RR95|D2RR95_HALTV Uncharacterized protein OS=Haloterrigena turkmenica (strain ATCC 51198 / DSM 5511 / JCM 9101 / NCIMB 13204 / VKM B-1734 / 4k) OX=543526 GN=Htur_3629 PE=4 SV=1
MM1 pKa = 7.94ALRR4 pKa = 11.84KK5 pKa = 9.81LLGSKK10 pKa = 8.54ATRR13 pKa = 11.84SLTVVSVLSDD23 pKa = 3.23AKK25 pKa = 10.62RR26 pKa = 11.84AFNRR30 pKa = 11.84GNRR33 pKa = 11.84TRR35 pKa = 11.84GLLLVGVAVLAWKK48 pKa = 7.96WTVLGMAAQGIVKK61 pKa = 10.0LLRR64 pKa = 11.84RR65 pKa = 11.84GGSAASSPAA74 pKa = 3.1
MM1 pKa = 7.94ALRR4 pKa = 11.84KK5 pKa = 9.81LLGSKK10 pKa = 8.54ATRR13 pKa = 11.84SLTVVSVLSDD23 pKa = 3.23AKK25 pKa = 10.62RR26 pKa = 11.84AFNRR30 pKa = 11.84GNRR33 pKa = 11.84TRR35 pKa = 11.84GLLLVGVAVLAWKK48 pKa = 7.96WTVLGMAAQGIVKK61 pKa = 10.0LLRR64 pKa = 11.84RR65 pKa = 11.84GGSAASSPAA74 pKa = 3.1
Molecular weight: 7.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1473976 |
30 |
1969 |
288.3 |
31.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.565 ± 0.052 | 0.77 ± 0.012 |
8.632 ± 0.042 | 9.243 ± 0.052 |
3.231 ± 0.022 | 8.321 ± 0.037 |
1.984 ± 0.016 | 4.374 ± 0.027 |
1.713 ± 0.019 | 8.837 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.753 ± 0.015 | 2.384 ± 0.019 |
4.654 ± 0.025 | 2.428 ± 0.021 |
6.566 ± 0.039 | 5.616 ± 0.026 |
6.465 ± 0.025 | 8.558 ± 0.039 |
1.167 ± 0.015 | 2.739 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
