
Camel associated drosmacovirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Drosmacovirus
Average proteome isoelectric point is 5.0
Get precalculated fractions of proteins
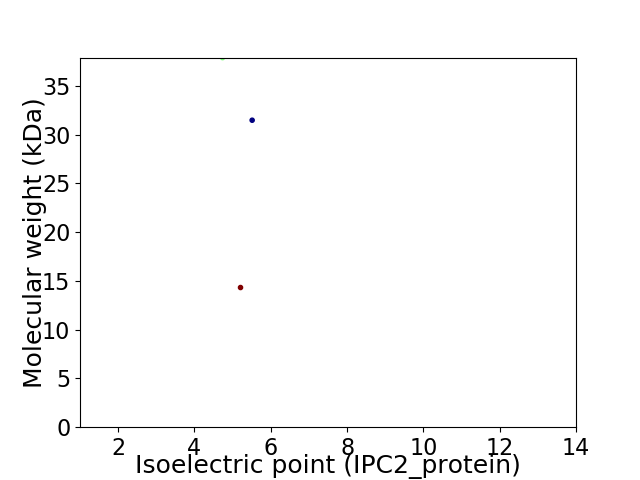
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A1ENW6|A0A0A1ENW6_9VIRU Putative replicase protein OS=Camel associated drosmacovirus 2 OX=2169877 GN=rep PE=4 SV=1
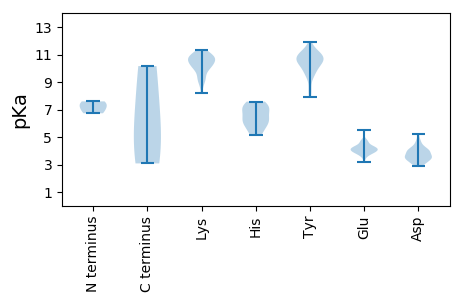
MM1 pKa = 7.23HH2 pKa = 7.58AEE4 pKa = 4.14DD5 pKa = 4.11TSVPMDD11 pKa = 4.74AGDD14 pKa = 3.47TDD16 pKa = 3.93TAGGIDD22 pKa = 4.26MVTVRR27 pKa = 11.84YY28 pKa = 8.33TEE30 pKa = 4.31TYY32 pKa = 10.54DD33 pKa = 3.25LCTEE37 pKa = 4.02VGKK40 pKa = 10.88LGIIGIHH47 pKa = 5.94TPTLSNLEE55 pKa = 3.92YY56 pKa = 10.44LYY58 pKa = 10.74RR59 pKa = 11.84GLMMNHH65 pKa = 6.98RR66 pKa = 11.84YY67 pKa = 8.63VHH69 pKa = 7.09LDD71 pKa = 3.0GCDD74 pKa = 3.41VVIACASTLPADD86 pKa = 4.05PLQVGTEE93 pKa = 4.04AGDD96 pKa = 3.58VAPQDD101 pKa = 3.55MFNPILYY108 pKa = 9.77KK109 pKa = 10.6AVSNDD114 pKa = 2.93SFNTVINRR122 pKa = 11.84VTVGSSVLDD131 pKa = 3.89DD132 pKa = 5.2EE133 pKa = 4.67ITGGSVRR140 pKa = 11.84KK141 pKa = 10.43VGDD144 pKa = 3.41AFADD148 pKa = 4.29LPQLANAPEE157 pKa = 3.79RR158 pKa = 11.84VYY160 pKa = 11.19YY161 pKa = 10.66GLLADD166 pKa = 4.1PDD168 pKa = 3.85GWKK171 pKa = 10.41KK172 pKa = 11.05AMPQAGLEE180 pKa = 4.02MRR182 pKa = 11.84GLYY185 pKa = 9.9PIVHH189 pKa = 7.19TIYY192 pKa = 9.65STYY195 pKa = 10.36GQQTSPGSNPDD206 pKa = 3.58AQSLPYY212 pKa = 10.61YY213 pKa = 10.42SDD215 pKa = 3.13AGQRR219 pKa = 11.84GSANRR224 pKa = 11.84EE225 pKa = 3.64YY226 pKa = 10.59YY227 pKa = 10.75LRR229 pKa = 11.84GNSVRR234 pKa = 11.84LPRR237 pKa = 11.84LPLCAGTQSSGASGDD252 pKa = 3.54QLPVVTIPTTYY263 pKa = 9.97VACLVMPPAKK273 pKa = 9.91LHH275 pKa = 5.29KK276 pKa = 9.36FYY278 pKa = 11.35YY279 pKa = 9.75RR280 pKa = 11.84MRR282 pKa = 11.84VTWTITFSEE291 pKa = 4.76VVPLTEE297 pKa = 4.1WEE299 pKa = 4.25NPTRR303 pKa = 11.84LWDD306 pKa = 3.36VGTYY310 pKa = 9.38VYY312 pKa = 10.84KK313 pKa = 10.42SDD315 pKa = 4.04YY316 pKa = 10.93DD317 pKa = 3.96EE318 pKa = 3.87QSKK321 pKa = 10.94RR322 pKa = 11.84MDD324 pKa = 3.58TTEE327 pKa = 3.95STVDD331 pKa = 3.34AKK333 pKa = 10.7DD334 pKa = 3.39TTLEE338 pKa = 4.28RR339 pKa = 11.84IMTAGKK345 pKa = 10.16
MM1 pKa = 7.23HH2 pKa = 7.58AEE4 pKa = 4.14DD5 pKa = 4.11TSVPMDD11 pKa = 4.74AGDD14 pKa = 3.47TDD16 pKa = 3.93TAGGIDD22 pKa = 4.26MVTVRR27 pKa = 11.84YY28 pKa = 8.33TEE30 pKa = 4.31TYY32 pKa = 10.54DD33 pKa = 3.25LCTEE37 pKa = 4.02VGKK40 pKa = 10.88LGIIGIHH47 pKa = 5.94TPTLSNLEE55 pKa = 3.92YY56 pKa = 10.44LYY58 pKa = 10.74RR59 pKa = 11.84GLMMNHH65 pKa = 6.98RR66 pKa = 11.84YY67 pKa = 8.63VHH69 pKa = 7.09LDD71 pKa = 3.0GCDD74 pKa = 3.41VVIACASTLPADD86 pKa = 4.05PLQVGTEE93 pKa = 4.04AGDD96 pKa = 3.58VAPQDD101 pKa = 3.55MFNPILYY108 pKa = 9.77KK109 pKa = 10.6AVSNDD114 pKa = 2.93SFNTVINRR122 pKa = 11.84VTVGSSVLDD131 pKa = 3.89DD132 pKa = 5.2EE133 pKa = 4.67ITGGSVRR140 pKa = 11.84KK141 pKa = 10.43VGDD144 pKa = 3.41AFADD148 pKa = 4.29LPQLANAPEE157 pKa = 3.79RR158 pKa = 11.84VYY160 pKa = 11.19YY161 pKa = 10.66GLLADD166 pKa = 4.1PDD168 pKa = 3.85GWKK171 pKa = 10.41KK172 pKa = 11.05AMPQAGLEE180 pKa = 4.02MRR182 pKa = 11.84GLYY185 pKa = 9.9PIVHH189 pKa = 7.19TIYY192 pKa = 9.65STYY195 pKa = 10.36GQQTSPGSNPDD206 pKa = 3.58AQSLPYY212 pKa = 10.61YY213 pKa = 10.42SDD215 pKa = 3.13AGQRR219 pKa = 11.84GSANRR224 pKa = 11.84EE225 pKa = 3.64YY226 pKa = 10.59YY227 pKa = 10.75LRR229 pKa = 11.84GNSVRR234 pKa = 11.84LPRR237 pKa = 11.84LPLCAGTQSSGASGDD252 pKa = 3.54QLPVVTIPTTYY263 pKa = 9.97VACLVMPPAKK273 pKa = 9.91LHH275 pKa = 5.29KK276 pKa = 9.36FYY278 pKa = 11.35YY279 pKa = 9.75RR280 pKa = 11.84MRR282 pKa = 11.84VTWTITFSEE291 pKa = 4.76VVPLTEE297 pKa = 4.1WEE299 pKa = 4.25NPTRR303 pKa = 11.84LWDD306 pKa = 3.36VGTYY310 pKa = 9.38VYY312 pKa = 10.84KK313 pKa = 10.42SDD315 pKa = 4.04YY316 pKa = 10.93DD317 pKa = 3.96EE318 pKa = 3.87QSKK321 pKa = 10.94RR322 pKa = 11.84MDD324 pKa = 3.58TTEE327 pKa = 3.95STVDD331 pKa = 3.34AKK333 pKa = 10.7DD334 pKa = 3.39TTLEE338 pKa = 4.28RR339 pKa = 11.84IMTAGKK345 pKa = 10.16
Molecular weight: 37.89 kDa
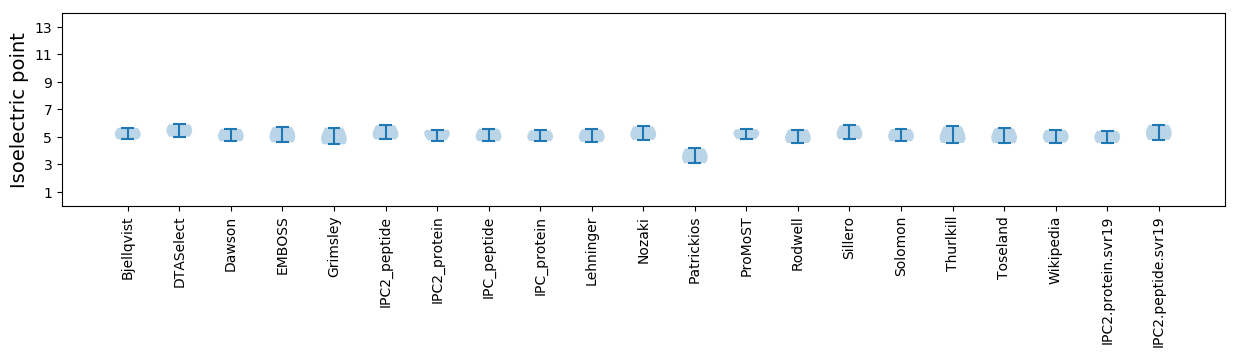
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1ENW6|A0A0A1ENW6_9VIRU Putative replicase protein OS=Camel associated drosmacovirus 2 OX=2169877 GN=rep PE=4 SV=1
MM1 pKa = 7.59TNPTWFCGTIWKK13 pKa = 10.06DD14 pKa = 3.07KK15 pKa = 10.28YY16 pKa = 11.03SGSKK20 pKa = 9.26EE21 pKa = 3.76QLAEE25 pKa = 3.88WFRR28 pKa = 11.84LHH30 pKa = 6.38HH31 pKa = 6.4AKK33 pKa = 10.5EE34 pKa = 4.21GVIGEE39 pKa = 4.59EE40 pKa = 3.92ISPTSGKK47 pKa = 9.11IHH49 pKa = 5.16YY50 pKa = 7.91QFRR53 pKa = 11.84IHH55 pKa = 7.37LDD57 pKa = 2.9RR58 pKa = 11.84GEE60 pKa = 4.22TLKK63 pKa = 10.77GWQEE67 pKa = 3.89LIKK70 pKa = 10.75AVGHH74 pKa = 5.86VEE76 pKa = 3.91VCQEE80 pKa = 4.19KK81 pKa = 10.73KK82 pKa = 9.46FTDD85 pKa = 3.82YY86 pKa = 11.12EE87 pKa = 4.37KK88 pKa = 11.18KK89 pKa = 10.35DD90 pKa = 3.77GNWIEE95 pKa = 4.09WPEE98 pKa = 4.07SYY100 pKa = 10.38LGKK103 pKa = 10.26FKK105 pKa = 10.82EE106 pKa = 4.19MPLLFWQEE114 pKa = 3.7QLLEE118 pKa = 4.06QWNEE122 pKa = 3.5QDD124 pKa = 3.66DD125 pKa = 4.12RR126 pKa = 11.84QILFVKK132 pKa = 10.0DD133 pKa = 3.5VKK135 pKa = 11.04GGNGKK140 pKa = 8.19STFGKK145 pKa = 10.05IMEE148 pKa = 4.33ARR150 pKa = 11.84GLEE153 pKa = 4.4EE154 pKa = 4.01VCPVCSDD161 pKa = 4.36DD162 pKa = 4.09YY163 pKa = 11.9NDD165 pKa = 3.55YY166 pKa = 11.02TSFCLEE172 pKa = 4.28YY173 pKa = 9.46PAKK176 pKa = 10.54GYY178 pKa = 10.51IFDD181 pKa = 3.94IPRR184 pKa = 11.84ASSIKK189 pKa = 10.11RR190 pKa = 11.84RR191 pKa = 11.84TALWSGIEE199 pKa = 3.95QIKK202 pKa = 10.96NGLLYY207 pKa = 10.24EE208 pKa = 4.18KK209 pKa = 9.82RR210 pKa = 11.84YY211 pKa = 10.05KK212 pKa = 9.1PRR214 pKa = 11.84KK215 pKa = 8.82VWIEE219 pKa = 3.72PPKK222 pKa = 10.98VIVFTNEE229 pKa = 4.67DD230 pKa = 3.82EE231 pKa = 4.73PPWDD235 pKa = 4.42LLSLDD240 pKa = 3.21RR241 pKa = 11.84WRR243 pKa = 11.84VFDD246 pKa = 3.48VEE248 pKa = 4.73QYY250 pKa = 11.23VNPITYY256 pKa = 10.24NIRR259 pKa = 11.84SHH261 pKa = 6.29WEE263 pKa = 3.2VDD265 pKa = 3.08
MM1 pKa = 7.59TNPTWFCGTIWKK13 pKa = 10.06DD14 pKa = 3.07KK15 pKa = 10.28YY16 pKa = 11.03SGSKK20 pKa = 9.26EE21 pKa = 3.76QLAEE25 pKa = 3.88WFRR28 pKa = 11.84LHH30 pKa = 6.38HH31 pKa = 6.4AKK33 pKa = 10.5EE34 pKa = 4.21GVIGEE39 pKa = 4.59EE40 pKa = 3.92ISPTSGKK47 pKa = 9.11IHH49 pKa = 5.16YY50 pKa = 7.91QFRR53 pKa = 11.84IHH55 pKa = 7.37LDD57 pKa = 2.9RR58 pKa = 11.84GEE60 pKa = 4.22TLKK63 pKa = 10.77GWQEE67 pKa = 3.89LIKK70 pKa = 10.75AVGHH74 pKa = 5.86VEE76 pKa = 3.91VCQEE80 pKa = 4.19KK81 pKa = 10.73KK82 pKa = 9.46FTDD85 pKa = 3.82YY86 pKa = 11.12EE87 pKa = 4.37KK88 pKa = 11.18KK89 pKa = 10.35DD90 pKa = 3.77GNWIEE95 pKa = 4.09WPEE98 pKa = 4.07SYY100 pKa = 10.38LGKK103 pKa = 10.26FKK105 pKa = 10.82EE106 pKa = 4.19MPLLFWQEE114 pKa = 3.7QLLEE118 pKa = 4.06QWNEE122 pKa = 3.5QDD124 pKa = 3.66DD125 pKa = 4.12RR126 pKa = 11.84QILFVKK132 pKa = 10.0DD133 pKa = 3.5VKK135 pKa = 11.04GGNGKK140 pKa = 8.19STFGKK145 pKa = 10.05IMEE148 pKa = 4.33ARR150 pKa = 11.84GLEE153 pKa = 4.4EE154 pKa = 4.01VCPVCSDD161 pKa = 4.36DD162 pKa = 4.09YY163 pKa = 11.9NDD165 pKa = 3.55YY166 pKa = 11.02TSFCLEE172 pKa = 4.28YY173 pKa = 9.46PAKK176 pKa = 10.54GYY178 pKa = 10.51IFDD181 pKa = 3.94IPRR184 pKa = 11.84ASSIKK189 pKa = 10.11RR190 pKa = 11.84RR191 pKa = 11.84TALWSGIEE199 pKa = 3.95QIKK202 pKa = 10.96NGLLYY207 pKa = 10.24EE208 pKa = 4.18KK209 pKa = 9.82RR210 pKa = 11.84YY211 pKa = 10.05KK212 pKa = 9.1PRR214 pKa = 11.84KK215 pKa = 8.82VWIEE219 pKa = 3.72PPKK222 pKa = 10.98VIVFTNEE229 pKa = 4.67DD230 pKa = 3.82EE231 pKa = 4.73PPWDD235 pKa = 4.42LLSLDD240 pKa = 3.21RR241 pKa = 11.84WRR243 pKa = 11.84VFDD246 pKa = 3.48VEE248 pKa = 4.73QYY250 pKa = 11.23VNPITYY256 pKa = 10.24NIRR259 pKa = 11.84SHH261 pKa = 6.29WEE263 pKa = 3.2VDD265 pKa = 3.08
Molecular weight: 31.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
739 |
129 |
345 |
246.3 |
27.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.413 ± 1.279 | 1.353 ± 0.347 |
7.307 ± 0.548 | 6.225 ± 1.709 |
2.842 ± 0.879 | 8.525 ± 1.185 |
1.894 ± 0.159 | 5.007 ± 0.841 |
5.277 ± 1.656 | 7.848 ± 0.294 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.436 ± 0.665 | 3.383 ± 0.147 |
5.142 ± 0.883 | 3.518 ± 0.275 |
5.142 ± 0.269 | 6.089 ± 0.543 |
7.037 ± 1.57 | 7.037 ± 0.832 |
2.706 ± 1.043 | 5.819 ± 0.606 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
