
Stenotrophomonas phage PSH1
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Psecadovirus; Stenotrophomonas virus PSH1
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
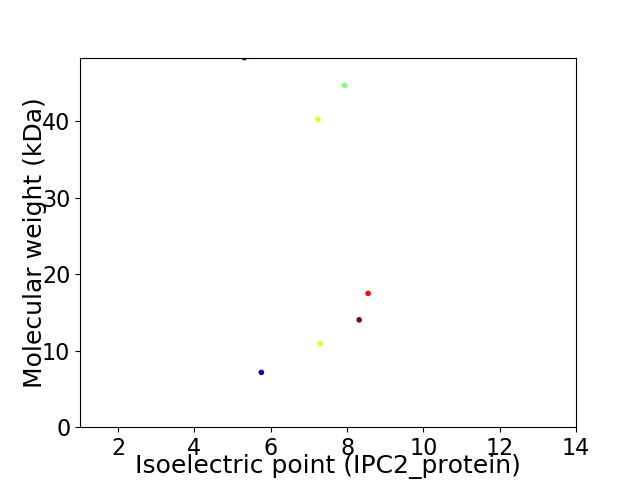
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B1NI81|B1NI81_9VIRU Zot domain-containing protein OS=Stenotrophomonas phage PSH1 OX=2011087 PE=4 SV=1
MM1 pKa = 7.55ARR3 pKa = 11.84TGWLIQLSLLVLLGVAPWSAHH24 pKa = 5.44AQAQQCANNSANCTQPQAYY43 pKa = 8.39AACRR47 pKa = 11.84AEE49 pKa = 4.12LSSYY53 pKa = 10.16IAIRR57 pKa = 11.84KK58 pKa = 9.27DD59 pKa = 3.24FANQNMEE66 pKa = 4.39CVPVGSGGRR75 pKa = 11.84TIQSKK80 pKa = 9.03FQYY83 pKa = 8.42KK84 pKa = 9.59TSANSAWFSGNYY96 pKa = 7.48RR97 pKa = 11.84TYY99 pKa = 9.86TWTKK103 pKa = 8.77GCDD106 pKa = 3.3VEE108 pKa = 4.32PTYY111 pKa = 10.87TGTGPWSSGGSAKK124 pKa = 10.53NGSIGCRR131 pKa = 11.84NGCDD135 pKa = 4.35GMWSTNADD143 pKa = 3.32ATKK146 pKa = 9.02TWTPTGDD153 pKa = 2.96ICPEE157 pKa = 3.97DD158 pKa = 3.49EE159 pKa = 5.71KK160 pKa = 11.5KK161 pKa = 10.81NCDD164 pKa = 3.32HH165 pKa = 6.79MGDD168 pKa = 3.59GYY170 pKa = 11.52YY171 pKa = 9.92WNSLLKK177 pKa = 10.55VCEE180 pKa = 4.19PPEE183 pKa = 4.5GKK185 pKa = 10.35CPGGTKK191 pKa = 10.06PNSLGQCAPEE201 pKa = 4.14PCPDD205 pKa = 4.35GMAQQPDD212 pKa = 4.32GTCKK216 pKa = 10.41KK217 pKa = 10.51KK218 pKa = 10.88EE219 pKa = 4.1NEE221 pKa = 4.21CPAGQVRR228 pKa = 11.84SPDD231 pKa = 3.64GRR233 pKa = 11.84CLPGDD238 pKa = 3.75GQCAKK243 pKa = 10.96GEE245 pKa = 4.1VRR247 pKa = 11.84GPDD250 pKa = 3.59GTCKK254 pKa = 10.1KK255 pKa = 10.75DD256 pKa = 3.2SDD258 pKa = 4.39NDD260 pKa = 4.09GQPDD264 pKa = 3.61EE265 pKa = 4.98PGEE268 pKa = 4.19KK269 pKa = 9.97EE270 pKa = 4.22SFAGGDD276 pKa = 3.95DD277 pKa = 3.52CSMPPSCSGSPILCGQARR295 pKa = 11.84IQWRR299 pKa = 11.84IDD301 pKa = 3.09CNTRR305 pKa = 11.84RR306 pKa = 11.84NRR308 pKa = 11.84NIAGGSCAAMPVCTGEE324 pKa = 3.99KK325 pKa = 9.52CDD327 pKa = 3.47AMEE330 pKa = 3.98YY331 pKa = 10.78AGLLMQWRR339 pKa = 11.84TACAAEE345 pKa = 4.21KK346 pKa = 10.43LASQGNNNGGDD357 pKa = 3.74GAQPEE362 pKa = 4.4WTKK365 pKa = 11.11VGGMSTDD372 pKa = 3.55PGLGASPDD380 pKa = 3.66DD381 pKa = 3.93TKK383 pKa = 11.61VLTVKK388 pKa = 10.75RR389 pKa = 11.84LGVEE393 pKa = 3.89QLDD396 pKa = 3.61QSGFGGGGSCIGFAVSGGSGIGSGFAQAMASPPDD430 pKa = 3.99FFCSYY435 pKa = 10.2IILIRR440 pKa = 11.84AVIILSATVTCAFILTSGGKK460 pKa = 8.72NN461 pKa = 3.0
MM1 pKa = 7.55ARR3 pKa = 11.84TGWLIQLSLLVLLGVAPWSAHH24 pKa = 5.44AQAQQCANNSANCTQPQAYY43 pKa = 8.39AACRR47 pKa = 11.84AEE49 pKa = 4.12LSSYY53 pKa = 10.16IAIRR57 pKa = 11.84KK58 pKa = 9.27DD59 pKa = 3.24FANQNMEE66 pKa = 4.39CVPVGSGGRR75 pKa = 11.84TIQSKK80 pKa = 9.03FQYY83 pKa = 8.42KK84 pKa = 9.59TSANSAWFSGNYY96 pKa = 7.48RR97 pKa = 11.84TYY99 pKa = 9.86TWTKK103 pKa = 8.77GCDD106 pKa = 3.3VEE108 pKa = 4.32PTYY111 pKa = 10.87TGTGPWSSGGSAKK124 pKa = 10.53NGSIGCRR131 pKa = 11.84NGCDD135 pKa = 4.35GMWSTNADD143 pKa = 3.32ATKK146 pKa = 9.02TWTPTGDD153 pKa = 2.96ICPEE157 pKa = 3.97DD158 pKa = 3.49EE159 pKa = 5.71KK160 pKa = 11.5KK161 pKa = 10.81NCDD164 pKa = 3.32HH165 pKa = 6.79MGDD168 pKa = 3.59GYY170 pKa = 11.52YY171 pKa = 9.92WNSLLKK177 pKa = 10.55VCEE180 pKa = 4.19PPEE183 pKa = 4.5GKK185 pKa = 10.35CPGGTKK191 pKa = 10.06PNSLGQCAPEE201 pKa = 4.14PCPDD205 pKa = 4.35GMAQQPDD212 pKa = 4.32GTCKK216 pKa = 10.41KK217 pKa = 10.51KK218 pKa = 10.88EE219 pKa = 4.1NEE221 pKa = 4.21CPAGQVRR228 pKa = 11.84SPDD231 pKa = 3.64GRR233 pKa = 11.84CLPGDD238 pKa = 3.75GQCAKK243 pKa = 10.96GEE245 pKa = 4.1VRR247 pKa = 11.84GPDD250 pKa = 3.59GTCKK254 pKa = 10.1KK255 pKa = 10.75DD256 pKa = 3.2SDD258 pKa = 4.39NDD260 pKa = 4.09GQPDD264 pKa = 3.61EE265 pKa = 4.98PGEE268 pKa = 4.19KK269 pKa = 9.97EE270 pKa = 4.22SFAGGDD276 pKa = 3.95DD277 pKa = 3.52CSMPPSCSGSPILCGQARR295 pKa = 11.84IQWRR299 pKa = 11.84IDD301 pKa = 3.09CNTRR305 pKa = 11.84RR306 pKa = 11.84NRR308 pKa = 11.84NIAGGSCAAMPVCTGEE324 pKa = 3.99KK325 pKa = 9.52CDD327 pKa = 3.47AMEE330 pKa = 3.98YY331 pKa = 10.78AGLLMQWRR339 pKa = 11.84TACAAEE345 pKa = 4.21KK346 pKa = 10.43LASQGNNNGGDD357 pKa = 3.74GAQPEE362 pKa = 4.4WTKK365 pKa = 11.11VGGMSTDD372 pKa = 3.55PGLGASPDD380 pKa = 3.66DD381 pKa = 3.93TKK383 pKa = 11.61VLTVKK388 pKa = 10.75RR389 pKa = 11.84LGVEE393 pKa = 3.89QLDD396 pKa = 3.61QSGFGGGGSCIGFAVSGGSGIGSGFAQAMASPPDD430 pKa = 3.99FFCSYY435 pKa = 10.2IILIRR440 pKa = 11.84AVIILSATVTCAFILTSGGKK460 pKa = 8.72NN461 pKa = 3.0
Molecular weight: 48.29 kDa
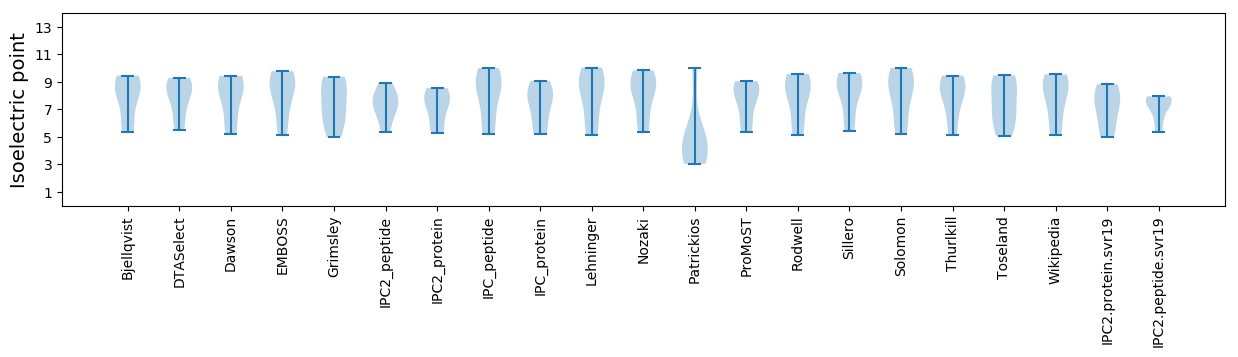
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B1NI83|B1NI83_9VIRU Uncharacterized protein OS=Stenotrophomonas phage PSH1 OX=2011087 PE=4 SV=1
MM1 pKa = 8.19IEE3 pKa = 4.44FDD5 pKa = 3.21PHH7 pKa = 7.65HH8 pKa = 7.12RR9 pKa = 11.84IDD11 pKa = 5.1LTGPWAGFSFLGDD24 pKa = 3.29RR25 pKa = 11.84LITPEE30 pKa = 3.89GRR32 pKa = 11.84EE33 pKa = 4.23LLPEE37 pKa = 4.29DD38 pKa = 4.58LAWLSLTACQAQEE51 pKa = 3.72WRR53 pKa = 11.84RR54 pKa = 11.84MMEE57 pKa = 4.05AARR60 pKa = 11.84SVPSIDD66 pKa = 3.21SSRR69 pKa = 11.84NVSNRR74 pKa = 11.84NAGIRR79 pKa = 11.84HH80 pKa = 5.87HH81 pKa = 7.06PATVVNLRR89 pKa = 11.84DD90 pKa = 3.5VVSRR94 pKa = 11.84RR95 pKa = 11.84KK96 pKa = 9.6QRR98 pKa = 11.84SAVAIAGPDD107 pKa = 3.64AEE109 pKa = 4.94PPAAGLPIQGPMPRR123 pKa = 11.84QRR125 pKa = 11.84MM126 pKa = 3.56
MM1 pKa = 8.19IEE3 pKa = 4.44FDD5 pKa = 3.21PHH7 pKa = 7.65HH8 pKa = 7.12RR9 pKa = 11.84IDD11 pKa = 5.1LTGPWAGFSFLGDD24 pKa = 3.29RR25 pKa = 11.84LITPEE30 pKa = 3.89GRR32 pKa = 11.84EE33 pKa = 4.23LLPEE37 pKa = 4.29DD38 pKa = 4.58LAWLSLTACQAQEE51 pKa = 3.72WRR53 pKa = 11.84RR54 pKa = 11.84MMEE57 pKa = 4.05AARR60 pKa = 11.84SVPSIDD66 pKa = 3.21SSRR69 pKa = 11.84NVSNRR74 pKa = 11.84NAGIRR79 pKa = 11.84HH80 pKa = 5.87HH81 pKa = 7.06PATVVNLRR89 pKa = 11.84DD90 pKa = 3.5VVSRR94 pKa = 11.84RR95 pKa = 11.84KK96 pKa = 9.6QRR98 pKa = 11.84SAVAIAGPDD107 pKa = 3.64AEE109 pKa = 4.94PPAAGLPIQGPMPRR123 pKa = 11.84QRR125 pKa = 11.84MM126 pKa = 3.56
Molecular weight: 14.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1683 |
68 |
461 |
240.4 |
26.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.23 ± 0.744 | 3.268 ± 0.855 |
5.585 ± 0.447 | 4.813 ± 0.458 |
3.209 ± 0.483 | 9.745 ± 1.095 |
1.485 ± 0.453 | 3.922 ± 0.319 |
4.694 ± 0.603 | 7.071 ± 0.678 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.436 ± 0.328 | 3.03 ± 0.505 |
6.239 ± 0.851 | 4.04 ± 0.461 |
6.358 ± 0.819 | 6.001 ± 0.67 |
5.823 ± 0.733 | 5.823 ± 0.744 |
2.139 ± 0.294 | 3.09 ± 0.649 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
