
Melon chlorotic spot virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Tenuivirus; Melon tenuivirus
Average proteome isoelectric point is 5.78
Get precalculated fractions of proteins
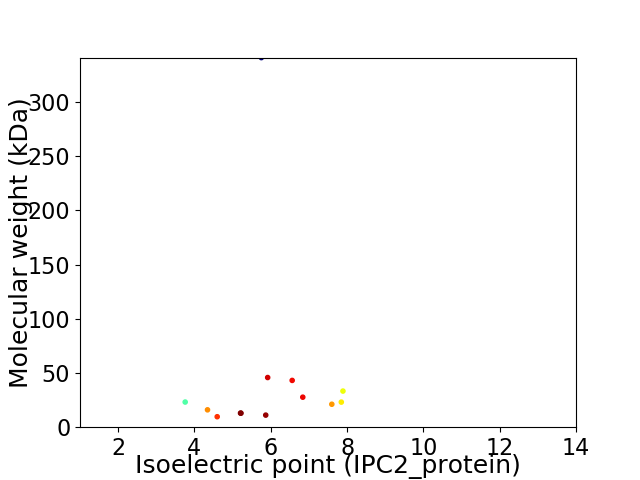
Virtual 2D-PAGE plot for 13 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G1Z2Z8|A0A3G1Z2Z8_9VIRU Uncharacterized protein OS=Melon chlorotic spot virus OX=2479459 PE=4 SV=1
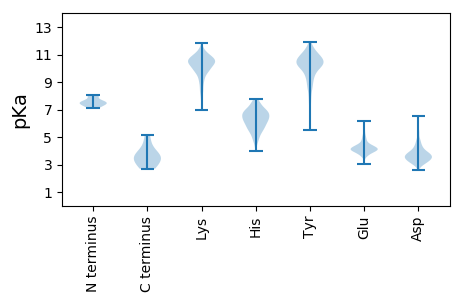
MM1 pKa = 8.05DD2 pKa = 6.49LIEE5 pKa = 4.71VDD7 pKa = 3.56NLYY10 pKa = 11.13DD11 pKa = 4.83LIEE14 pKa = 4.1YY15 pKa = 10.23CKK17 pKa = 10.84AKK19 pKa = 10.44GLNSSCLVPMVLHH32 pKa = 5.62NTGIHH37 pKa = 6.31VDD39 pKa = 3.1INNNDD44 pKa = 2.96FRR46 pKa = 11.84LDD48 pKa = 3.43HH49 pKa = 6.63MDD51 pKa = 3.22WKK53 pKa = 11.16LEE55 pKa = 3.97EE56 pKa = 4.9PDD58 pKa = 3.85PNIEE62 pKa = 3.92INLNLVFEE70 pKa = 5.8DD71 pKa = 3.13IRR73 pKa = 11.84EE74 pKa = 4.35TISMAEE80 pKa = 4.06DD81 pKa = 3.55EE82 pKa = 5.28AWYY85 pKa = 10.29QPDD88 pKa = 4.12EE89 pKa = 4.01ICYY92 pKa = 10.31GIRR95 pKa = 11.84DD96 pKa = 4.11LDD98 pKa = 4.77QLVLWCQKK106 pKa = 9.49EE107 pKa = 4.11RR108 pKa = 11.84TFFPEE113 pKa = 3.85LLSLVFDD120 pKa = 4.22SMPIYY125 pKa = 10.28ISFMEE130 pKa = 4.48PLVFLDD136 pKa = 2.79VDD138 pKa = 4.07EE139 pKa = 5.05PVSIPLYY146 pKa = 10.35HH147 pKa = 6.73SFAEE151 pKa = 4.82MIDD154 pKa = 3.55QVIQDD159 pKa = 3.32IQMEE163 pKa = 4.46IEE165 pKa = 4.15CKK167 pKa = 9.84IYY169 pKa = 10.5HH170 pKa = 7.49DD171 pKa = 5.58DD172 pKa = 3.69YY173 pKa = 11.92DD174 pKa = 4.39SYY176 pKa = 11.72FYY178 pKa = 11.46LLDD181 pKa = 4.11SDD183 pKa = 3.99DD184 pKa = 4.44ARR186 pKa = 11.84WEE188 pKa = 4.42GEE190 pKa = 4.05DD191 pKa = 3.83YY192 pKa = 11.04LYY194 pKa = 11.11DD195 pKa = 3.43
MM1 pKa = 8.05DD2 pKa = 6.49LIEE5 pKa = 4.71VDD7 pKa = 3.56NLYY10 pKa = 11.13DD11 pKa = 4.83LIEE14 pKa = 4.1YY15 pKa = 10.23CKK17 pKa = 10.84AKK19 pKa = 10.44GLNSSCLVPMVLHH32 pKa = 5.62NTGIHH37 pKa = 6.31VDD39 pKa = 3.1INNNDD44 pKa = 2.96FRR46 pKa = 11.84LDD48 pKa = 3.43HH49 pKa = 6.63MDD51 pKa = 3.22WKK53 pKa = 11.16LEE55 pKa = 3.97EE56 pKa = 4.9PDD58 pKa = 3.85PNIEE62 pKa = 3.92INLNLVFEE70 pKa = 5.8DD71 pKa = 3.13IRR73 pKa = 11.84EE74 pKa = 4.35TISMAEE80 pKa = 4.06DD81 pKa = 3.55EE82 pKa = 5.28AWYY85 pKa = 10.29QPDD88 pKa = 4.12EE89 pKa = 4.01ICYY92 pKa = 10.31GIRR95 pKa = 11.84DD96 pKa = 4.11LDD98 pKa = 4.77QLVLWCQKK106 pKa = 9.49EE107 pKa = 4.11RR108 pKa = 11.84TFFPEE113 pKa = 3.85LLSLVFDD120 pKa = 4.22SMPIYY125 pKa = 10.28ISFMEE130 pKa = 4.48PLVFLDD136 pKa = 2.79VDD138 pKa = 4.07EE139 pKa = 5.05PVSIPLYY146 pKa = 10.35HH147 pKa = 6.73SFAEE151 pKa = 4.82MIDD154 pKa = 3.55QVIQDD159 pKa = 3.32IQMEE163 pKa = 4.46IEE165 pKa = 4.15CKK167 pKa = 9.84IYY169 pKa = 10.5HH170 pKa = 7.49DD171 pKa = 5.58DD172 pKa = 3.69YY173 pKa = 11.92DD174 pKa = 4.39SYY176 pKa = 11.72FYY178 pKa = 11.46LLDD181 pKa = 4.11SDD183 pKa = 3.99DD184 pKa = 4.44ARR186 pKa = 11.84WEE188 pKa = 4.42GEE190 pKa = 4.05DD191 pKa = 3.83YY192 pKa = 11.04LYY194 pKa = 11.11DD195 pKa = 3.43
Molecular weight: 23.21 kDa
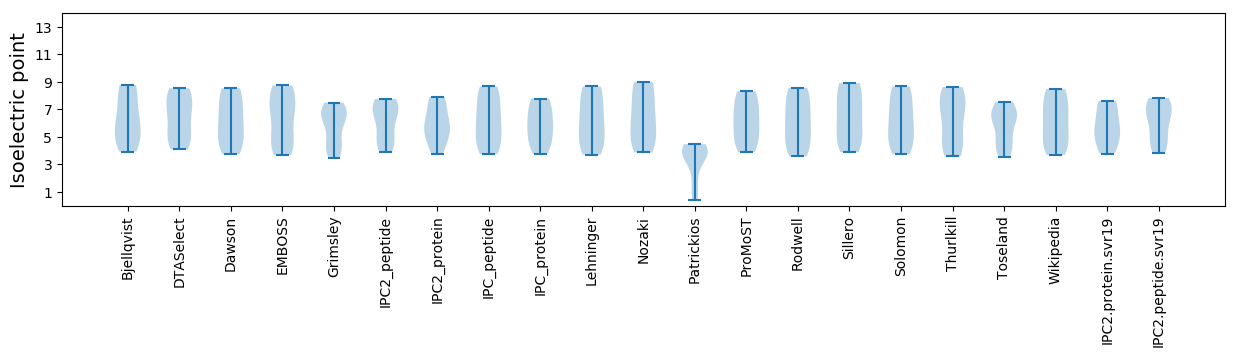
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G1Z1G8|A0A3G1Z1G8_9VIRU Uncharacterized protein OS=Melon chlorotic spot virus OX=2479459 PE=4 SV=1
MM1 pKa = 7.54APVNSLADD9 pKa = 3.34VQGILNQIKK18 pKa = 10.41GVDD21 pKa = 3.9LSDD24 pKa = 3.84SSIEE28 pKa = 3.9QTLSGFVYY36 pKa = 10.46AGFNAVEE43 pKa = 4.12MWKK46 pKa = 9.78MIGSTPGFDD55 pKa = 3.97PKK57 pKa = 10.89DD58 pKa = 3.29IALLICACLQKK69 pKa = 11.02GYY71 pKa = 10.05GLKK74 pKa = 10.54KK75 pKa = 10.35FSTKK79 pKa = 9.41VKK81 pKa = 8.8QTAAKK86 pKa = 8.41NTIDD90 pKa = 5.49AIVLKK95 pKa = 10.59YY96 pKa = 10.49NIKK99 pKa = 8.88PTSKK103 pKa = 10.56DD104 pKa = 3.2SSDD107 pKa = 3.31PTLQRR112 pKa = 11.84IVSTSGIIAFNCYY125 pKa = 7.08QHH127 pKa = 6.45VLSKK131 pKa = 11.03GRR133 pKa = 11.84LNLAVPAASLGLKK146 pKa = 10.31CPDD149 pKa = 3.1VVGCSFINSIIGTVTEE165 pKa = 4.13TSVWEE170 pKa = 3.69PVLIVNEE177 pKa = 3.9YY178 pKa = 10.41LQAQVTMKK186 pKa = 10.38TMSEE190 pKa = 4.15EE191 pKa = 3.82NKK193 pKa = 9.73KK194 pKa = 9.97AQNITTVAEE203 pKa = 4.08AMLKK207 pKa = 10.07NRR209 pKa = 11.84AFSEE213 pKa = 4.4AARR216 pKa = 11.84SSPIVPSGDD225 pKa = 3.09DD226 pKa = 3.68RR227 pKa = 11.84MVLLNYY233 pKa = 9.66FRR235 pKa = 11.84GTNYY239 pKa = 10.05NFSPVSRR246 pKa = 11.84IPEE249 pKa = 4.41EE250 pKa = 3.98ISPGVYY256 pKa = 10.11DD257 pKa = 5.14DD258 pKa = 5.33LKK260 pKa = 10.94LIHH263 pKa = 6.49KK264 pKa = 7.79TLFTPEE270 pKa = 4.1FRR272 pKa = 11.84VQYY275 pKa = 9.86QAIGKK280 pKa = 9.31IGALMKK286 pKa = 10.59SCTQAEE292 pKa = 4.74FPSKK296 pKa = 10.17LTVSYY301 pKa = 11.06FNMAA305 pKa = 3.42
MM1 pKa = 7.54APVNSLADD9 pKa = 3.34VQGILNQIKK18 pKa = 10.41GVDD21 pKa = 3.9LSDD24 pKa = 3.84SSIEE28 pKa = 3.9QTLSGFVYY36 pKa = 10.46AGFNAVEE43 pKa = 4.12MWKK46 pKa = 9.78MIGSTPGFDD55 pKa = 3.97PKK57 pKa = 10.89DD58 pKa = 3.29IALLICACLQKK69 pKa = 11.02GYY71 pKa = 10.05GLKK74 pKa = 10.54KK75 pKa = 10.35FSTKK79 pKa = 9.41VKK81 pKa = 8.8QTAAKK86 pKa = 8.41NTIDD90 pKa = 5.49AIVLKK95 pKa = 10.59YY96 pKa = 10.49NIKK99 pKa = 8.88PTSKK103 pKa = 10.56DD104 pKa = 3.2SSDD107 pKa = 3.31PTLQRR112 pKa = 11.84IVSTSGIIAFNCYY125 pKa = 7.08QHH127 pKa = 6.45VLSKK131 pKa = 11.03GRR133 pKa = 11.84LNLAVPAASLGLKK146 pKa = 10.31CPDD149 pKa = 3.1VVGCSFINSIIGTVTEE165 pKa = 4.13TSVWEE170 pKa = 3.69PVLIVNEE177 pKa = 3.9YY178 pKa = 10.41LQAQVTMKK186 pKa = 10.38TMSEE190 pKa = 4.15EE191 pKa = 3.82NKK193 pKa = 9.73KK194 pKa = 9.97AQNITTVAEE203 pKa = 4.08AMLKK207 pKa = 10.07NRR209 pKa = 11.84AFSEE213 pKa = 4.4AARR216 pKa = 11.84SSPIVPSGDD225 pKa = 3.09DD226 pKa = 3.68RR227 pKa = 11.84MVLLNYY233 pKa = 9.66FRR235 pKa = 11.84GTNYY239 pKa = 10.05NFSPVSRR246 pKa = 11.84IPEE249 pKa = 4.41EE250 pKa = 3.98ISPGVYY256 pKa = 10.11DD257 pKa = 5.14DD258 pKa = 5.33LKK260 pKa = 10.94LIHH263 pKa = 6.49KK264 pKa = 7.79TLFTPEE270 pKa = 4.1FRR272 pKa = 11.84VQYY275 pKa = 9.86QAIGKK280 pKa = 9.31IGALMKK286 pKa = 10.59SCTQAEE292 pKa = 4.74FPSKK296 pKa = 10.17LTVSYY301 pKa = 11.06FNMAA305 pKa = 3.42
Molecular weight: 33.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5406 |
84 |
2943 |
415.8 |
47.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.033 ± 0.45 | 1.942 ± 0.469 |
6.03 ± 0.565 | 6.4 ± 0.409 |
4.532 ± 0.194 | 4.107 ± 0.396 |
1.961 ± 0.251 | 8.195 ± 0.401 |
6.641 ± 0.475 | 10.266 ± 0.495 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.108 ± 0.153 | 5.882 ± 0.295 |
3.7 ± 0.217 | 3.108 ± 0.215 |
4.477 ± 0.415 | 8.269 ± 0.319 |
5.771 ± 0.521 | 5.956 ± 0.33 |
0.999 ± 0.13 | 4.624 ± 0.264 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
