
Circovirus-like genome DHCV-3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.08
Get precalculated fractions of proteins
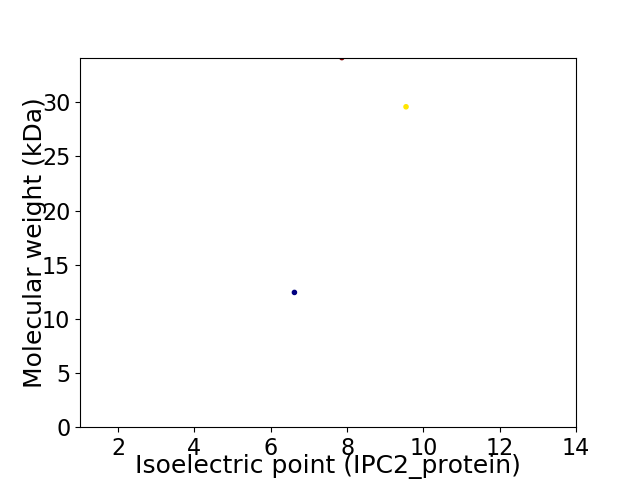
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A190WHE3|A0A190WHE3_9CIRC Uncharacterized protein OS=Circovirus-like genome DHCV-3 OX=1788452 PE=4 SV=1
MM1 pKa = 7.94AEE3 pKa = 4.14VNYY6 pKa = 9.81HH7 pKa = 5.82APKK10 pKa = 10.07NLHH13 pKa = 7.05LIPIGVPYY21 pKa = 11.01VSMLSIHH28 pKa = 6.33FLVAPLSYY36 pKa = 10.55NNSMHH41 pKa = 7.09LGGFHH46 pKa = 7.45LLFLYY51 pKa = 10.85VPDD54 pKa = 4.62LVTISSFSKK63 pKa = 10.7NFSHH67 pKa = 6.71SQNIHH72 pKa = 4.65NTVNALFLFPLPLYY86 pKa = 10.23QSQHH90 pKa = 5.36TDD92 pKa = 2.84TLQIIPGIPCNHH104 pKa = 6.29HH105 pKa = 6.74RR106 pKa = 11.84YY107 pKa = 10.04NYY109 pKa = 10.66
MM1 pKa = 7.94AEE3 pKa = 4.14VNYY6 pKa = 9.81HH7 pKa = 5.82APKK10 pKa = 10.07NLHH13 pKa = 7.05LIPIGVPYY21 pKa = 11.01VSMLSIHH28 pKa = 6.33FLVAPLSYY36 pKa = 10.55NNSMHH41 pKa = 7.09LGGFHH46 pKa = 7.45LLFLYY51 pKa = 10.85VPDD54 pKa = 4.62LVTISSFSKK63 pKa = 10.7NFSHH67 pKa = 6.71SQNIHH72 pKa = 4.65NTVNALFLFPLPLYY86 pKa = 10.23QSQHH90 pKa = 5.36TDD92 pKa = 2.84TLQIIPGIPCNHH104 pKa = 6.29HH105 pKa = 6.74RR106 pKa = 11.84YY107 pKa = 10.04NYY109 pKa = 10.66
Molecular weight: 12.44 kDa
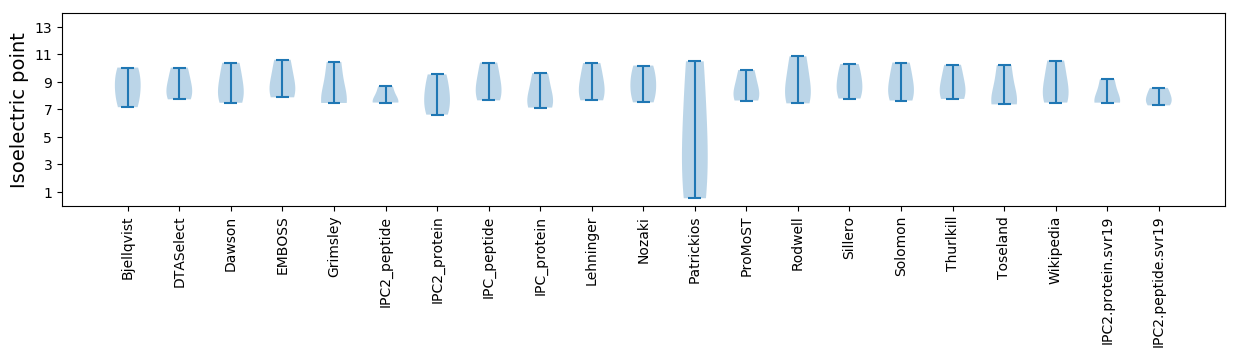
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A190WHG0|A0A190WHG0_9CIRC ATP-dependent helicase Rep OS=Circovirus-like genome DHCV-3 OX=1788452 PE=3 SV=1
MM1 pKa = 7.65AYY3 pKa = 9.68KK4 pKa = 10.52RR5 pKa = 11.84NRR7 pKa = 11.84RR8 pKa = 11.84GGRR11 pKa = 11.84KK12 pKa = 7.56TNVKK16 pKa = 9.92KK17 pKa = 10.57AVRR20 pKa = 11.84ANNRR24 pKa = 11.84ARR26 pKa = 11.84QYY28 pKa = 10.23NAPAKK33 pKa = 8.67VASVVSLAKK42 pKa = 10.24SVNRR46 pKa = 11.84LKK48 pKa = 10.35TKK50 pKa = 8.21MRR52 pKa = 11.84RR53 pKa = 11.84EE54 pKa = 4.0TEE56 pKa = 3.93LKK58 pKa = 10.13EE59 pKa = 4.16FEE61 pKa = 4.46SGLLSTTVGQVNINNTGMYY80 pKa = 9.71IDD82 pKa = 5.79DD83 pKa = 5.53LDD85 pKa = 5.21LMDD88 pKa = 4.26IAEE91 pKa = 4.55GASDD95 pKa = 3.39KK96 pKa = 11.0TRR98 pKa = 11.84IGKK101 pKa = 9.44KK102 pKa = 10.25VMLKK106 pKa = 10.07GVQVRR111 pKa = 11.84LQFSHH116 pKa = 6.4QSNTSSASNYY126 pKa = 9.34IIEE129 pKa = 4.38VYY131 pKa = 8.89KK132 pKa = 9.88TQDD135 pKa = 2.95FGSTLTAIRR144 pKa = 11.84DD145 pKa = 3.49ALYY148 pKa = 10.87NVDD151 pKa = 4.74SISGVIDD158 pKa = 3.42YY159 pKa = 11.22SSTHH163 pKa = 5.87KK164 pKa = 11.12NEE166 pKa = 3.83AKK168 pKa = 10.16SWLKK172 pKa = 9.87RR173 pKa = 11.84VAYY176 pKa = 9.47KK177 pKa = 10.57RR178 pKa = 11.84IRR180 pKa = 11.84LPVDD184 pKa = 3.44TFTGVNMVKK193 pKa = 10.14DD194 pKa = 3.53VKK196 pKa = 10.71MFIKK200 pKa = 10.29QNQEE204 pKa = 3.16LTYY207 pKa = 9.91TLGSTQVPQNYY218 pKa = 9.54RR219 pKa = 11.84YY220 pKa = 9.6IICIRR225 pKa = 11.84SSVGNNNATTASTLTTVVHH244 pKa = 5.6TTALTGAIVRR254 pKa = 11.84LVSKK258 pKa = 10.32CWYY261 pKa = 8.38TDD263 pKa = 2.91NN264 pKa = 5.04
MM1 pKa = 7.65AYY3 pKa = 9.68KK4 pKa = 10.52RR5 pKa = 11.84NRR7 pKa = 11.84RR8 pKa = 11.84GGRR11 pKa = 11.84KK12 pKa = 7.56TNVKK16 pKa = 9.92KK17 pKa = 10.57AVRR20 pKa = 11.84ANNRR24 pKa = 11.84ARR26 pKa = 11.84QYY28 pKa = 10.23NAPAKK33 pKa = 8.67VASVVSLAKK42 pKa = 10.24SVNRR46 pKa = 11.84LKK48 pKa = 10.35TKK50 pKa = 8.21MRR52 pKa = 11.84RR53 pKa = 11.84EE54 pKa = 4.0TEE56 pKa = 3.93LKK58 pKa = 10.13EE59 pKa = 4.16FEE61 pKa = 4.46SGLLSTTVGQVNINNTGMYY80 pKa = 9.71IDD82 pKa = 5.79DD83 pKa = 5.53LDD85 pKa = 5.21LMDD88 pKa = 4.26IAEE91 pKa = 4.55GASDD95 pKa = 3.39KK96 pKa = 11.0TRR98 pKa = 11.84IGKK101 pKa = 9.44KK102 pKa = 10.25VMLKK106 pKa = 10.07GVQVRR111 pKa = 11.84LQFSHH116 pKa = 6.4QSNTSSASNYY126 pKa = 9.34IIEE129 pKa = 4.38VYY131 pKa = 8.89KK132 pKa = 9.88TQDD135 pKa = 2.95FGSTLTAIRR144 pKa = 11.84DD145 pKa = 3.49ALYY148 pKa = 10.87NVDD151 pKa = 4.74SISGVIDD158 pKa = 3.42YY159 pKa = 11.22SSTHH163 pKa = 5.87KK164 pKa = 11.12NEE166 pKa = 3.83AKK168 pKa = 10.16SWLKK172 pKa = 9.87RR173 pKa = 11.84VAYY176 pKa = 9.47KK177 pKa = 10.57RR178 pKa = 11.84IRR180 pKa = 11.84LPVDD184 pKa = 3.44TFTGVNMVKK193 pKa = 10.14DD194 pKa = 3.53VKK196 pKa = 10.71MFIKK200 pKa = 10.29QNQEE204 pKa = 3.16LTYY207 pKa = 9.91TLGSTQVPQNYY218 pKa = 9.54RR219 pKa = 11.84YY220 pKa = 9.6IICIRR225 pKa = 11.84SSVGNNNATTASTLTTVVHH244 pKa = 5.6TTALTGAIVRR254 pKa = 11.84LVSKK258 pKa = 10.32CWYY261 pKa = 8.38TDD263 pKa = 2.91NN264 pKa = 5.04
Molecular weight: 29.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
663 |
109 |
290 |
221.0 |
25.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.581 ± 0.831 | 1.357 ± 0.419 |
4.525 ± 0.772 | 5.128 ± 2.073 |
2.866 ± 0.913 | 5.43 ± 0.546 |
2.866 ± 1.619 | 5.581 ± 0.538 |
7.692 ± 1.555 | 7.994 ± 1.478 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.866 ± 0.141 | 6.637 ± 1.094 |
3.318 ± 1.394 | 3.771 ± 0.026 |
5.882 ± 1.272 | 7.24 ± 1.017 |
6.637 ± 1.634 | 7.391 ± 1.093 |
1.659 ± 0.854 | 5.581 ± 0.528 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
