
Camellia chlorotic dwarf-associated virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Citlodavirus
Average proteome isoelectric point is 7.25
Get precalculated fractions of proteins
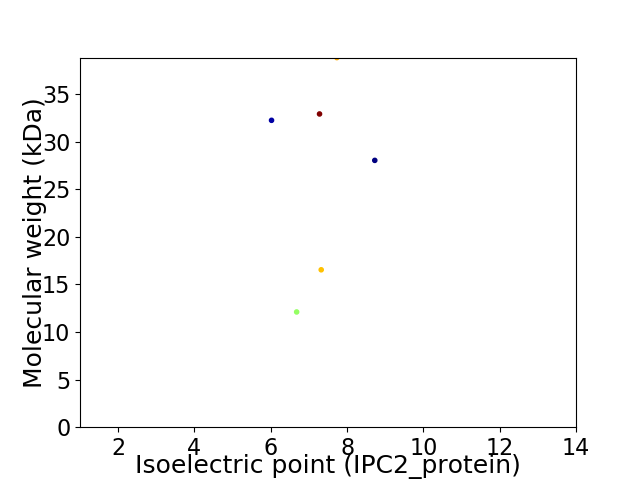
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1JHH9|A0A2P1JHH9_9GEMI Replication-associated protein OS=Camellia chlorotic dwarf-associated virus OX=2122733 PE=3 SV=1
MM1 pKa = 7.84APTQKK6 pKa = 10.01QFRR9 pKa = 11.84FASKK13 pKa = 10.83NIFLTFAQCNLLMATLLDD31 pKa = 3.95HH32 pKa = 7.34LKK34 pKa = 10.65HH35 pKa = 6.2LLSNLDD41 pKa = 3.29IIYY44 pKa = 10.2ISVCSEE50 pKa = 3.34LHH52 pKa = 6.63ASGDD56 pKa = 3.78PHH58 pKa = 7.9LHH60 pKa = 7.01AMVQLKK66 pKa = 10.4KK67 pKa = 10.19PLQTRR72 pKa = 11.84NPRR75 pKa = 11.84FFDD78 pKa = 3.71YY79 pKa = 11.27NSDD82 pKa = 3.55RR83 pKa = 11.84NNFHH87 pKa = 7.37PSFEE91 pKa = 4.17PLRR94 pKa = 11.84NPRR97 pKa = 11.84ASKK100 pKa = 10.69SYY102 pKa = 9.6IQKK105 pKa = 10.14DD106 pKa = 3.67GNFIEE111 pKa = 4.83EE112 pKa = 5.47GEE114 pKa = 4.2FQSNGKK120 pKa = 9.47SPHH123 pKa = 5.92KK124 pKa = 9.5PANKK128 pKa = 8.39VWQEE132 pKa = 3.8ILQKK136 pKa = 10.07STDD139 pKa = 3.33EE140 pKa = 3.92NDD142 pKa = 3.29FYY144 pKa = 11.58KK145 pKa = 9.79QVKK148 pKa = 5.61MHH150 pKa = 6.77RR151 pKa = 11.84PADD154 pKa = 3.89FVLRR158 pKa = 11.84WPAISSFAKK167 pKa = 10.13DD168 pKa = 3.04HH169 pKa = 5.91YY170 pKa = 10.67NRR172 pKa = 11.84NFIPYY177 pKa = 8.37TPQFEE182 pKa = 4.64NFVNLPDD189 pKa = 5.76NIRR192 pKa = 11.84EE193 pKa = 4.03WANNNVLCVSKK204 pKa = 9.69TFVQYY209 pKa = 10.57DD210 pKa = 3.69LCYY213 pKa = 9.29EE214 pKa = 4.2CSKK217 pKa = 11.05KK218 pKa = 10.97AMFEE222 pKa = 4.28TEE224 pKa = 4.28CPISLLHH231 pKa = 7.05NFYY234 pKa = 10.81CDD236 pKa = 3.21NCRR239 pKa = 11.84SPSLDD244 pKa = 4.09LIVHH248 pKa = 6.22EE249 pKa = 5.38ASTSVDD255 pKa = 3.04QAEE258 pKa = 4.49QEE260 pKa = 4.26RR261 pKa = 11.84HH262 pKa = 5.87PGQDD266 pKa = 2.99PSDD269 pKa = 4.21YY270 pKa = 10.49IIISQTPP277 pKa = 2.89
MM1 pKa = 7.84APTQKK6 pKa = 10.01QFRR9 pKa = 11.84FASKK13 pKa = 10.83NIFLTFAQCNLLMATLLDD31 pKa = 3.95HH32 pKa = 7.34LKK34 pKa = 10.65HH35 pKa = 6.2LLSNLDD41 pKa = 3.29IIYY44 pKa = 10.2ISVCSEE50 pKa = 3.34LHH52 pKa = 6.63ASGDD56 pKa = 3.78PHH58 pKa = 7.9LHH60 pKa = 7.01AMVQLKK66 pKa = 10.4KK67 pKa = 10.19PLQTRR72 pKa = 11.84NPRR75 pKa = 11.84FFDD78 pKa = 3.71YY79 pKa = 11.27NSDD82 pKa = 3.55RR83 pKa = 11.84NNFHH87 pKa = 7.37PSFEE91 pKa = 4.17PLRR94 pKa = 11.84NPRR97 pKa = 11.84ASKK100 pKa = 10.69SYY102 pKa = 9.6IQKK105 pKa = 10.14DD106 pKa = 3.67GNFIEE111 pKa = 4.83EE112 pKa = 5.47GEE114 pKa = 4.2FQSNGKK120 pKa = 9.47SPHH123 pKa = 5.92KK124 pKa = 9.5PANKK128 pKa = 8.39VWQEE132 pKa = 3.8ILQKK136 pKa = 10.07STDD139 pKa = 3.33EE140 pKa = 3.92NDD142 pKa = 3.29FYY144 pKa = 11.58KK145 pKa = 9.79QVKK148 pKa = 5.61MHH150 pKa = 6.77RR151 pKa = 11.84PADD154 pKa = 3.89FVLRR158 pKa = 11.84WPAISSFAKK167 pKa = 10.13DD168 pKa = 3.04HH169 pKa = 5.91YY170 pKa = 10.67NRR172 pKa = 11.84NFIPYY177 pKa = 8.37TPQFEE182 pKa = 4.64NFVNLPDD189 pKa = 5.76NIRR192 pKa = 11.84EE193 pKa = 4.03WANNNVLCVSKK204 pKa = 9.69TFVQYY209 pKa = 10.57DD210 pKa = 3.69LCYY213 pKa = 9.29EE214 pKa = 4.2CSKK217 pKa = 11.05KK218 pKa = 10.97AMFEE222 pKa = 4.28TEE224 pKa = 4.28CPISLLHH231 pKa = 7.05NFYY234 pKa = 10.81CDD236 pKa = 3.21NCRR239 pKa = 11.84SPSLDD244 pKa = 4.09LIVHH248 pKa = 6.22EE249 pKa = 5.38ASTSVDD255 pKa = 3.04QAEE258 pKa = 4.49QEE260 pKa = 4.26RR261 pKa = 11.84HH262 pKa = 5.87PGQDD266 pKa = 2.99PSDD269 pKa = 4.21YY270 pKa = 10.49IIISQTPP277 pKa = 2.89
Molecular weight: 32.24 kDa
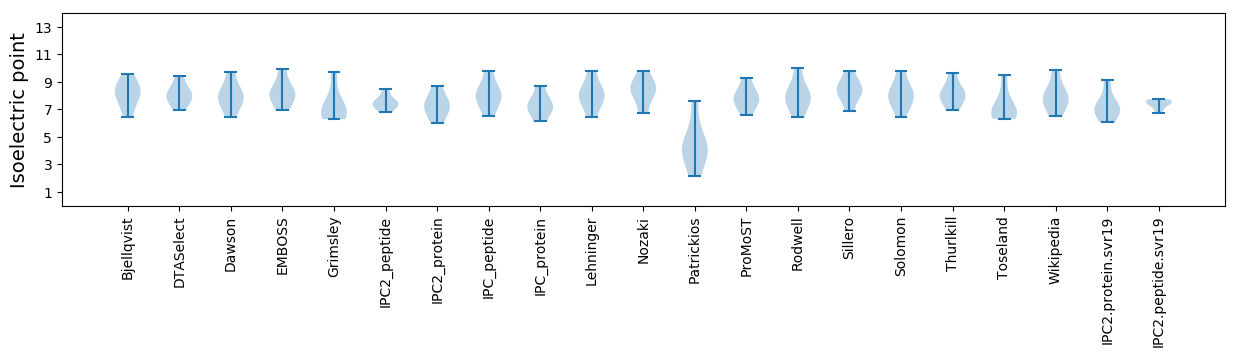
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1JHH5|A0A2P1JHH5_9GEMI Uncharacterized protein OS=Camellia chlorotic dwarf-associated virus OX=2122733 PE=4 SV=1
MM1 pKa = 7.29ATTRR5 pKa = 11.84SGRR8 pKa = 11.84VSGTVTDD15 pKa = 4.9RR16 pKa = 11.84LPMWRR21 pKa = 11.84SRR23 pKa = 11.84RR24 pKa = 11.84KK25 pKa = 9.15RR26 pKa = 11.84SARR29 pKa = 11.84SRR31 pKa = 11.84GSATSAARR39 pKa = 11.84RR40 pKa = 11.84SRR42 pKa = 11.84SAGVSIRR49 pKa = 11.84RR50 pKa = 11.84TGWKK54 pKa = 9.97GKK56 pKa = 10.55VPDD59 pKa = 4.31GCKK62 pKa = 10.42GPCKK66 pKa = 9.24THH68 pKa = 6.71SVDD71 pKa = 4.78LIKK74 pKa = 9.83TVYY77 pKa = 10.19HH78 pKa = 7.28DD79 pKa = 3.5GRR81 pKa = 11.84GAGFIGNIDD90 pKa = 3.5KK91 pKa = 11.55GEE93 pKa = 4.22DD94 pKa = 2.9LGNRR98 pKa = 11.84DD99 pKa = 2.97GRR101 pKa = 11.84RR102 pKa = 11.84IRR104 pKa = 11.84ITKK107 pKa = 9.35VLLRR111 pKa = 11.84GKK113 pKa = 9.79VWLDD117 pKa = 3.12VTNAALPGSNVCKK130 pKa = 10.37VWVIKK135 pKa = 10.1DD136 pKa = 3.54RR137 pKa = 11.84RR138 pKa = 11.84PGNEE142 pKa = 3.14IVNFTSLMDD151 pKa = 3.74MTDD154 pKa = 4.11SEE156 pKa = 4.67PMSALIKK163 pKa = 9.58MDD165 pKa = 3.17YY166 pKa = 9.9RR167 pKa = 11.84DD168 pKa = 3.57RR169 pKa = 11.84FIVLRR174 pKa = 11.84DD175 pKa = 3.52MQFDD179 pKa = 3.48LHH181 pKa = 6.62GGKK184 pKa = 10.13DD185 pKa = 3.6FRR187 pKa = 11.84VDD189 pKa = 3.41EE190 pKa = 4.52ASMDD194 pKa = 3.41EE195 pKa = 4.43MISVNCEE202 pKa = 3.72SLFSHH207 pKa = 6.7EE208 pKa = 5.12DD209 pKa = 3.22EE210 pKa = 5.63GNLSHH215 pKa = 7.05CLEE218 pKa = 4.43NGILVYY224 pKa = 8.39FACSDD229 pKa = 3.72PRR231 pKa = 11.84NAMQITAQCRR241 pKa = 11.84VYY243 pKa = 10.97FFDD246 pKa = 3.69STVNN250 pKa = 3.3
MM1 pKa = 7.29ATTRR5 pKa = 11.84SGRR8 pKa = 11.84VSGTVTDD15 pKa = 4.9RR16 pKa = 11.84LPMWRR21 pKa = 11.84SRR23 pKa = 11.84RR24 pKa = 11.84KK25 pKa = 9.15RR26 pKa = 11.84SARR29 pKa = 11.84SRR31 pKa = 11.84GSATSAARR39 pKa = 11.84RR40 pKa = 11.84SRR42 pKa = 11.84SAGVSIRR49 pKa = 11.84RR50 pKa = 11.84TGWKK54 pKa = 9.97GKK56 pKa = 10.55VPDD59 pKa = 4.31GCKK62 pKa = 10.42GPCKK66 pKa = 9.24THH68 pKa = 6.71SVDD71 pKa = 4.78LIKK74 pKa = 9.83TVYY77 pKa = 10.19HH78 pKa = 7.28DD79 pKa = 3.5GRR81 pKa = 11.84GAGFIGNIDD90 pKa = 3.5KK91 pKa = 11.55GEE93 pKa = 4.22DD94 pKa = 2.9LGNRR98 pKa = 11.84DD99 pKa = 2.97GRR101 pKa = 11.84RR102 pKa = 11.84IRR104 pKa = 11.84ITKK107 pKa = 9.35VLLRR111 pKa = 11.84GKK113 pKa = 9.79VWLDD117 pKa = 3.12VTNAALPGSNVCKK130 pKa = 10.37VWVIKK135 pKa = 10.1DD136 pKa = 3.54RR137 pKa = 11.84RR138 pKa = 11.84PGNEE142 pKa = 3.14IVNFTSLMDD151 pKa = 3.74MTDD154 pKa = 4.11SEE156 pKa = 4.67PMSALIKK163 pKa = 9.58MDD165 pKa = 3.17YY166 pKa = 9.9RR167 pKa = 11.84DD168 pKa = 3.57RR169 pKa = 11.84FIVLRR174 pKa = 11.84DD175 pKa = 3.52MQFDD179 pKa = 3.48LHH181 pKa = 6.62GGKK184 pKa = 10.13DD185 pKa = 3.6FRR187 pKa = 11.84VDD189 pKa = 3.41EE190 pKa = 4.52ASMDD194 pKa = 3.41EE195 pKa = 4.43MISVNCEE202 pKa = 3.72SLFSHH207 pKa = 6.7EE208 pKa = 5.12DD209 pKa = 3.22EE210 pKa = 5.63GNLSHH215 pKa = 7.05CLEE218 pKa = 4.43NGILVYY224 pKa = 8.39FACSDD229 pKa = 3.72PRR231 pKa = 11.84NAMQITAQCRR241 pKa = 11.84VYY243 pKa = 10.97FFDD246 pKa = 3.69STVNN250 pKa = 3.3
Molecular weight: 28.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1408 |
108 |
333 |
234.7 |
26.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.185 ± 0.412 | 2.983 ± 0.723 |
5.895 ± 0.636 | 5.398 ± 0.828 |
4.972 ± 0.602 | 5.611 ± 1.365 |
2.699 ± 0.601 | 5.398 ± 0.218 |
6.392 ± 0.412 | 7.528 ± 0.389 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.912 ± 0.343 | 5.824 ± 1.011 |
5.611 ± 0.678 | 3.196 ± 0.726 |
6.179 ± 1.02 | 8.594 ± 0.848 |
3.693 ± 0.701 | 6.676 ± 0.993 |
1.705 ± 0.252 | 3.551 ± 0.396 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
