
Batrachochytrium salamandrivorans
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Chytridiomycota; Chytridiomycota incertae sedis; Chytridiomycetes; Rhizophydiales; Rhizophydiales incertae sedis; Batrachochytrium
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
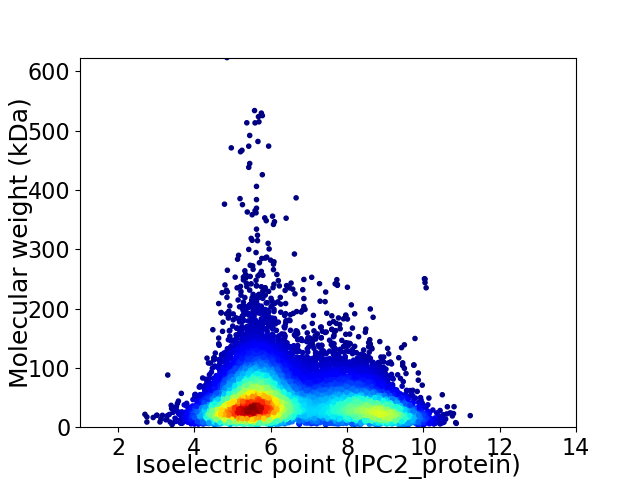
Virtual 2D-PAGE plot for 11933 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
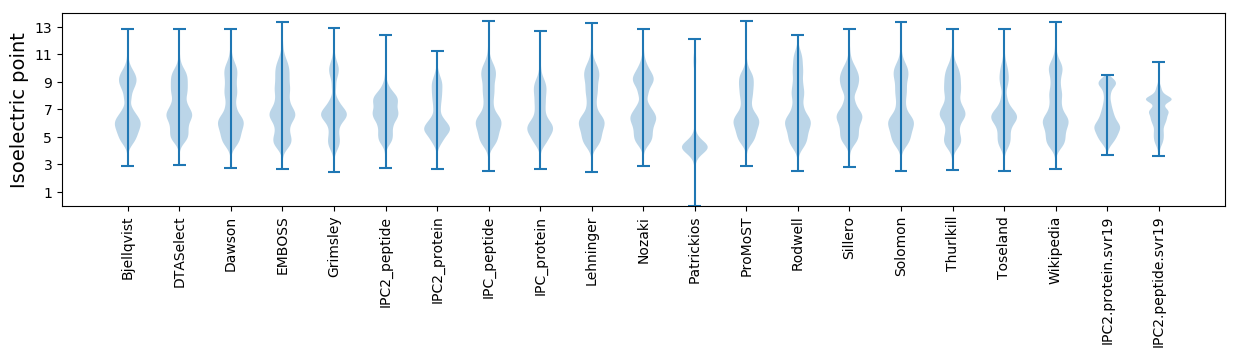
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S8VZV9|A0A1S8VZV9_9FUNG Uncharacterized protein OS=Batrachochytrium salamandrivorans OX=1357716 GN=BSLG_02834 PE=4 SV=1
MM1 pKa = 7.67EE2 pKa = 5.37GLDD5 pKa = 5.31RR6 pKa = 11.84PDD8 pKa = 4.33LLDD11 pKa = 3.44GRR13 pKa = 11.84TDD15 pKa = 3.0IGSTEE20 pKa = 4.15GEE22 pKa = 4.35DD23 pKa = 4.21DD24 pKa = 3.67VSADD28 pKa = 3.87SEE30 pKa = 4.4LADD33 pKa = 3.78TLAQIAAIDD42 pKa = 3.88EE43 pKa = 4.42LLAVASSAQSSDD55 pKa = 2.76LTYY58 pKa = 10.72PPTEE62 pKa = 4.1PTSNTITDD70 pKa = 3.86DD71 pKa = 3.62PAGISGLEE79 pKa = 3.84EE80 pKa = 4.81LIVARR85 pKa = 11.84NQLVEE90 pKa = 4.17LADD93 pKa = 4.01LQRR96 pKa = 11.84LLLSSAEE103 pKa = 4.25SVDD106 pKa = 3.73TSSTAAQHH114 pKa = 5.16VQSDD118 pKa = 4.33VAHH121 pKa = 6.79PSCSIDD127 pKa = 3.07IGEE130 pKa = 4.76SSTDD134 pKa = 3.22NNTDD138 pKa = 3.09NNADD142 pKa = 3.41NNTYY146 pKa = 11.06NNTYY150 pKa = 10.56NNTYY154 pKa = 10.46NNTVYY159 pKa = 10.66PSRR162 pKa = 11.84PTFAVGGLCCVVFSMSSSQQSFHH185 pKa = 7.17LLATITSLDD194 pKa = 3.55QPTNAQRR201 pKa = 11.84EE202 pKa = 4.17SDD204 pKa = 4.0ANLGSDD210 pKa = 4.85DD211 pKa = 4.08SDD213 pKa = 3.43IYY215 pKa = 9.93ATVMLLTPMTLEE227 pKa = 4.12TRR229 pKa = 11.84PCLDD233 pKa = 2.84IATCSRR239 pKa = 11.84EE240 pKa = 4.2CGRR243 pKa = 11.84SS244 pKa = 3.16
MM1 pKa = 7.67EE2 pKa = 5.37GLDD5 pKa = 5.31RR6 pKa = 11.84PDD8 pKa = 4.33LLDD11 pKa = 3.44GRR13 pKa = 11.84TDD15 pKa = 3.0IGSTEE20 pKa = 4.15GEE22 pKa = 4.35DD23 pKa = 4.21DD24 pKa = 3.67VSADD28 pKa = 3.87SEE30 pKa = 4.4LADD33 pKa = 3.78TLAQIAAIDD42 pKa = 3.88EE43 pKa = 4.42LLAVASSAQSSDD55 pKa = 2.76LTYY58 pKa = 10.72PPTEE62 pKa = 4.1PTSNTITDD70 pKa = 3.86DD71 pKa = 3.62PAGISGLEE79 pKa = 3.84EE80 pKa = 4.81LIVARR85 pKa = 11.84NQLVEE90 pKa = 4.17LADD93 pKa = 4.01LQRR96 pKa = 11.84LLLSSAEE103 pKa = 4.25SVDD106 pKa = 3.73TSSTAAQHH114 pKa = 5.16VQSDD118 pKa = 4.33VAHH121 pKa = 6.79PSCSIDD127 pKa = 3.07IGEE130 pKa = 4.76SSTDD134 pKa = 3.22NNTDD138 pKa = 3.09NNADD142 pKa = 3.41NNTYY146 pKa = 11.06NNTYY150 pKa = 10.56NNTYY154 pKa = 10.46NNTVYY159 pKa = 10.66PSRR162 pKa = 11.84PTFAVGGLCCVVFSMSSSQQSFHH185 pKa = 7.17LLATITSLDD194 pKa = 3.55QPTNAQRR201 pKa = 11.84EE202 pKa = 4.17SDD204 pKa = 4.0ANLGSDD210 pKa = 4.85DD211 pKa = 4.08SDD213 pKa = 3.43IYY215 pKa = 9.93ATVMLLTPMTLEE227 pKa = 4.12TRR229 pKa = 11.84PCLDD233 pKa = 2.84IATCSRR239 pKa = 11.84EE240 pKa = 4.2CGRR243 pKa = 11.84SS244 pKa = 3.16
Molecular weight: 25.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S8VUI4|A0A1S8VUI4_9FUNG Uncharacterized protein OS=Batrachochytrium salamandrivorans OX=1357716 GN=BSLG_04310 PE=4 SV=1
RR1 pKa = 7.9LQIPRR6 pKa = 11.84MLHH9 pKa = 5.53QRR11 pKa = 11.84LHH13 pKa = 5.79DD14 pKa = 3.32QYY16 pKa = 10.81MLHH19 pKa = 6.42QRR21 pKa = 11.84LQIPRR26 pKa = 11.84MLNQHH31 pKa = 6.62MISQRR36 pKa = 11.84MLNQHH41 pKa = 6.9MISQHH46 pKa = 5.49MLNQHH51 pKa = 6.63MISQRR56 pKa = 11.84MVNQRR61 pKa = 11.84MVNQRR66 pKa = 11.84MVNQRR71 pKa = 11.84MVNQRR76 pKa = 11.84MISQRR81 pKa = 11.84MISQRR86 pKa = 11.84MISQRR91 pKa = 11.84MISQRR96 pKa = 11.84MINQRR101 pKa = 11.84MVNQRR106 pKa = 11.84MVNQRR111 pKa = 11.84MINQRR116 pKa = 11.84MVNQRR121 pKa = 11.84MISQRR126 pKa = 11.84LQIPRR131 pKa = 11.84MLHH134 pKa = 5.01QRR136 pKa = 11.84LQIPRR141 pKa = 11.84MLHH144 pKa = 5.01QRR146 pKa = 11.84LQIPRR151 pKa = 11.84MLHH154 pKa = 3.95QQ155 pKa = 4.25
RR1 pKa = 7.9LQIPRR6 pKa = 11.84MLHH9 pKa = 5.53QRR11 pKa = 11.84LHH13 pKa = 5.79DD14 pKa = 3.32QYY16 pKa = 10.81MLHH19 pKa = 6.42QRR21 pKa = 11.84LQIPRR26 pKa = 11.84MLNQHH31 pKa = 6.62MISQRR36 pKa = 11.84MLNQHH41 pKa = 6.9MISQHH46 pKa = 5.49MLNQHH51 pKa = 6.63MISQRR56 pKa = 11.84MVNQRR61 pKa = 11.84MVNQRR66 pKa = 11.84MVNQRR71 pKa = 11.84MVNQRR76 pKa = 11.84MISQRR81 pKa = 11.84MISQRR86 pKa = 11.84MISQRR91 pKa = 11.84MISQRR96 pKa = 11.84MINQRR101 pKa = 11.84MVNQRR106 pKa = 11.84MVNQRR111 pKa = 11.84MINQRR116 pKa = 11.84MVNQRR121 pKa = 11.84MISQRR126 pKa = 11.84LQIPRR131 pKa = 11.84MLHH134 pKa = 5.01QRR136 pKa = 11.84LQIPRR141 pKa = 11.84MLHH144 pKa = 5.01QRR146 pKa = 11.84LQIPRR151 pKa = 11.84MLHH154 pKa = 3.95QQ155 pKa = 4.25
Molecular weight: 19.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5546497 |
30 |
5579 |
464.8 |
51.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.577 ± 0.021 | 1.402 ± 0.008 |
5.765 ± 0.017 | 5.261 ± 0.02 |
3.53 ± 0.017 | 5.665 ± 0.021 |
2.839 ± 0.013 | 5.739 ± 0.018 |
4.96 ± 0.022 | 9.43 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.452 ± 0.008 | 4.032 ± 0.011 |
5.39 ± 0.022 | 4.373 ± 0.022 |
5.217 ± 0.016 | 10.107 ± 0.031 |
6.541 ± 0.018 | 6.06 ± 0.017 |
0.994 ± 0.007 | 2.665 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
