
Tissierella sp. P1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Tissierellia; Tissierellales; Tissierellaceae; Tissierella; unclassified Tissierella
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
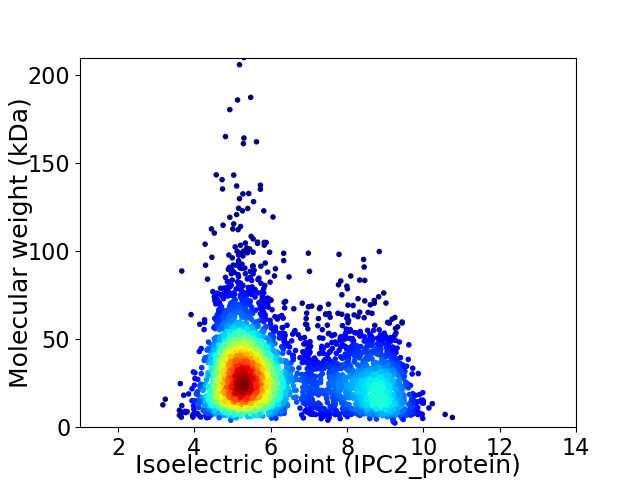
Virtual 2D-PAGE plot for 3981 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
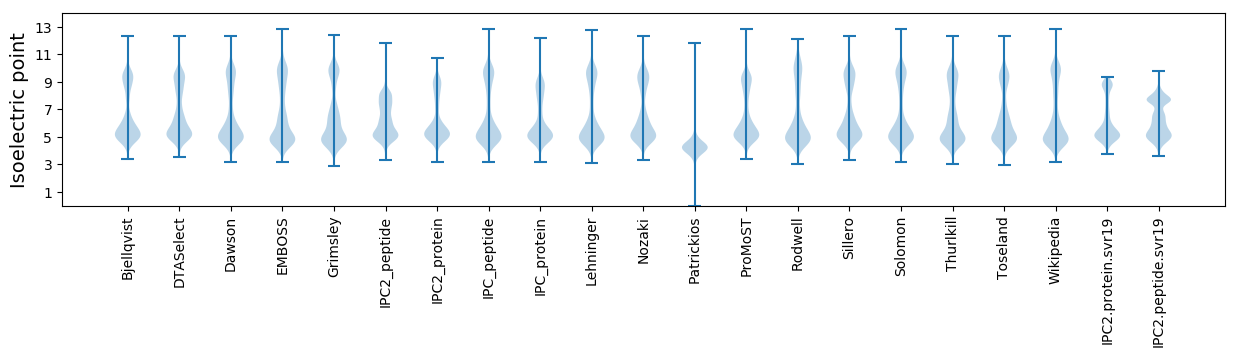
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A265Q181|A0A265Q181_9FIRM ATP-dependent zinc metalloprotease FtsH OS=Tissierella sp. P1 OX=1280483 GN=ftsH PE=3 SV=1
MM1 pKa = 7.15ATISGRR7 pKa = 11.84VVFDD11 pKa = 3.76RR12 pKa = 11.84DD13 pKa = 3.01RR14 pKa = 11.84SATINAGDD22 pKa = 3.69SGLANVPVVLQNTMTDD38 pKa = 2.84MRR40 pKa = 11.84LVVLTDD46 pKa = 3.24NNGNYY51 pKa = 10.53SFINVPNGSYY61 pKa = 10.43RR62 pKa = 11.84IVEE65 pKa = 4.4SYY67 pKa = 7.83GTPGGVATPGDD78 pKa = 4.12FNTAVVGPIPVGEE91 pKa = 4.68DD92 pKa = 3.47PPISFAPNPPSGSTNLDD109 pKa = 3.3SLTPDD114 pKa = 3.28TLLVTVSGFDD124 pKa = 3.36LTNQNFLDD132 pKa = 4.55GPVIYY137 pKa = 9.94TPIEE141 pKa = 4.5NILDD145 pKa = 3.84PCVSVSDD152 pKa = 3.61INLINVADD160 pKa = 3.97NGTFGSFPAGTPANTGAPVEE180 pKa = 4.5PYY182 pKa = 10.34PGVTPDD188 pKa = 3.43FTYY191 pKa = 11.09VLPDD195 pKa = 3.48PTDD198 pKa = 3.38YY199 pKa = 11.39TPFDD203 pKa = 3.46GEE205 pKa = 4.34YY206 pKa = 9.04TVQNIMTNALSNTIGAWWRR225 pKa = 11.84IADD228 pKa = 3.69HH229 pKa = 5.57TTGNEE234 pKa = 3.97TGRR237 pKa = 11.84MMVVNGFDD245 pKa = 3.25PGAVFFRR252 pKa = 11.84DD253 pKa = 3.66VVSVQPNTNYY263 pKa = 10.51LFSSWILNLFKK274 pKa = 10.74VTGYY278 pKa = 9.51PNPEE282 pKa = 3.96LGVRR286 pKa = 11.84ILDD289 pKa = 3.59QNGNPIYY296 pKa = 10.53SATLGAQIPVNTNTPEE312 pKa = 3.66WKK314 pKa = 10.34EE315 pKa = 3.48IGTVINSRR323 pKa = 11.84NNTSLTVEE331 pKa = 4.56FLSEE335 pKa = 4.04GPAAIGNDD343 pKa = 3.5YY344 pKa = 11.0AIDD347 pKa = 5.11DD348 pKa = 3.55ISLRR352 pKa = 11.84EE353 pKa = 3.79IQIPEE358 pKa = 3.96FTPVKK363 pKa = 9.7TVSTPTANIGDD374 pKa = 3.73IVTYY378 pKa = 9.77TVTLTNTCTSPLTNVFFQDD397 pKa = 3.39IVPNGLSFVPGSVTVNGSQDD417 pKa = 2.61ITLNPNVGFNVPDD430 pKa = 3.42IPGGSTATITFDD442 pKa = 3.56ALVDD446 pKa = 3.88NVPTPNPTLNTAMMSYY462 pKa = 10.28FYY464 pKa = 10.65TPVEE468 pKa = 4.05GGIEE472 pKa = 3.84NNYY475 pKa = 8.3TVNSNVVPLDD485 pKa = 3.19IAASADD491 pKa = 3.33VSVEE495 pKa = 4.17KK496 pKa = 10.17TSSPDD501 pKa = 3.41PVISGQVLQYY511 pKa = 10.12TIIVSNAGPSAAEE524 pKa = 3.63NVVLHH529 pKa = 6.83DD530 pKa = 5.39IIPPEE535 pKa = 3.57ITGAEE540 pKa = 4.19FSTDD544 pKa = 2.86GGATFSPWTGSYY556 pKa = 10.8DD557 pKa = 3.86IGTLLNGEE565 pKa = 4.31TRR567 pKa = 11.84TILIRR572 pKa = 11.84GTVSPTATGTITNTANVTSTTPDD595 pKa = 3.72PNPDD599 pKa = 3.48NNTSTTTTEE608 pKa = 3.83VNALADD614 pKa = 3.33IGVVKK619 pKa = 9.89TGTPNPVMSGEE630 pKa = 4.2TLTYY634 pKa = 10.02TIDD637 pKa = 3.34VSNFGPSDD645 pKa = 3.57AEE647 pKa = 4.09NVVLTDD653 pKa = 5.46AIPAEE658 pKa = 3.94ITGAEE663 pKa = 4.24FSTDD667 pKa = 2.89GGVTFSPWTGSYY679 pKa = 11.03NIGTLLNGEE688 pKa = 4.31TRR690 pKa = 11.84TILIRR695 pKa = 11.84GTVAPMAPGFITNTANVTSTTPDD718 pKa = 3.69PNPSNNTSTAVIEE731 pKa = 4.56VNQTTPVADD740 pKa = 3.29VGIFKK745 pKa = 10.64SVEE748 pKa = 4.03TNPIIPGGTVIYY760 pKa = 9.3PIEE763 pKa = 4.15VSNLGPSNAEE773 pKa = 3.7NVVLTDD779 pKa = 5.46AIPAEE784 pKa = 3.94ITGAEE789 pKa = 4.24FSTDD793 pKa = 2.58GGVTFNPWTGSYY805 pKa = 11.0NIGTLLNGEE814 pKa = 4.31TRR816 pKa = 11.84TILIRR821 pKa = 11.84GTVSPSATGIITNTANVTSTTPDD844 pKa = 3.58PNPP847 pKa = 3.47
MM1 pKa = 7.15ATISGRR7 pKa = 11.84VVFDD11 pKa = 3.76RR12 pKa = 11.84DD13 pKa = 3.01RR14 pKa = 11.84SATINAGDD22 pKa = 3.69SGLANVPVVLQNTMTDD38 pKa = 2.84MRR40 pKa = 11.84LVVLTDD46 pKa = 3.24NNGNYY51 pKa = 10.53SFINVPNGSYY61 pKa = 10.43RR62 pKa = 11.84IVEE65 pKa = 4.4SYY67 pKa = 7.83GTPGGVATPGDD78 pKa = 4.12FNTAVVGPIPVGEE91 pKa = 4.68DD92 pKa = 3.47PPISFAPNPPSGSTNLDD109 pKa = 3.3SLTPDD114 pKa = 3.28TLLVTVSGFDD124 pKa = 3.36LTNQNFLDD132 pKa = 4.55GPVIYY137 pKa = 9.94TPIEE141 pKa = 4.5NILDD145 pKa = 3.84PCVSVSDD152 pKa = 3.61INLINVADD160 pKa = 3.97NGTFGSFPAGTPANTGAPVEE180 pKa = 4.5PYY182 pKa = 10.34PGVTPDD188 pKa = 3.43FTYY191 pKa = 11.09VLPDD195 pKa = 3.48PTDD198 pKa = 3.38YY199 pKa = 11.39TPFDD203 pKa = 3.46GEE205 pKa = 4.34YY206 pKa = 9.04TVQNIMTNALSNTIGAWWRR225 pKa = 11.84IADD228 pKa = 3.69HH229 pKa = 5.57TTGNEE234 pKa = 3.97TGRR237 pKa = 11.84MMVVNGFDD245 pKa = 3.25PGAVFFRR252 pKa = 11.84DD253 pKa = 3.66VVSVQPNTNYY263 pKa = 10.51LFSSWILNLFKK274 pKa = 10.74VTGYY278 pKa = 9.51PNPEE282 pKa = 3.96LGVRR286 pKa = 11.84ILDD289 pKa = 3.59QNGNPIYY296 pKa = 10.53SATLGAQIPVNTNTPEE312 pKa = 3.66WKK314 pKa = 10.34EE315 pKa = 3.48IGTVINSRR323 pKa = 11.84NNTSLTVEE331 pKa = 4.56FLSEE335 pKa = 4.04GPAAIGNDD343 pKa = 3.5YY344 pKa = 11.0AIDD347 pKa = 5.11DD348 pKa = 3.55ISLRR352 pKa = 11.84EE353 pKa = 3.79IQIPEE358 pKa = 3.96FTPVKK363 pKa = 9.7TVSTPTANIGDD374 pKa = 3.73IVTYY378 pKa = 9.77TVTLTNTCTSPLTNVFFQDD397 pKa = 3.39IVPNGLSFVPGSVTVNGSQDD417 pKa = 2.61ITLNPNVGFNVPDD430 pKa = 3.42IPGGSTATITFDD442 pKa = 3.56ALVDD446 pKa = 3.88NVPTPNPTLNTAMMSYY462 pKa = 10.28FYY464 pKa = 10.65TPVEE468 pKa = 4.05GGIEE472 pKa = 3.84NNYY475 pKa = 8.3TVNSNVVPLDD485 pKa = 3.19IAASADD491 pKa = 3.33VSVEE495 pKa = 4.17KK496 pKa = 10.17TSSPDD501 pKa = 3.41PVISGQVLQYY511 pKa = 10.12TIIVSNAGPSAAEE524 pKa = 3.63NVVLHH529 pKa = 6.83DD530 pKa = 5.39IIPPEE535 pKa = 3.57ITGAEE540 pKa = 4.19FSTDD544 pKa = 2.86GGATFSPWTGSYY556 pKa = 10.8DD557 pKa = 3.86IGTLLNGEE565 pKa = 4.31TRR567 pKa = 11.84TILIRR572 pKa = 11.84GTVSPTATGTITNTANVTSTTPDD595 pKa = 3.72PNPDD599 pKa = 3.48NNTSTTTTEE608 pKa = 3.83VNALADD614 pKa = 3.33IGVVKK619 pKa = 9.89TGTPNPVMSGEE630 pKa = 4.2TLTYY634 pKa = 10.02TIDD637 pKa = 3.34VSNFGPSDD645 pKa = 3.57AEE647 pKa = 4.09NVVLTDD653 pKa = 5.46AIPAEE658 pKa = 3.94ITGAEE663 pKa = 4.24FSTDD667 pKa = 2.89GGVTFSPWTGSYY679 pKa = 11.03NIGTLLNGEE688 pKa = 4.31TRR690 pKa = 11.84TILIRR695 pKa = 11.84GTVAPMAPGFITNTANVTSTTPDD718 pKa = 3.69PNPSNNTSTAVIEE731 pKa = 4.56VNQTTPVADD740 pKa = 3.29VGIFKK745 pKa = 10.64SVEE748 pKa = 4.03TNPIIPGGTVIYY760 pKa = 9.3PIEE763 pKa = 4.15VSNLGPSNAEE773 pKa = 3.7NVVLTDD779 pKa = 5.46AIPAEE784 pKa = 3.94ITGAEE789 pKa = 4.24FSTDD793 pKa = 2.58GGVTFNPWTGSYY805 pKa = 11.0NIGTLLNGEE814 pKa = 4.31TRR816 pKa = 11.84TILIRR821 pKa = 11.84GTVSPSATGIITNTANVTSTTPDD844 pKa = 3.58PNPP847 pKa = 3.47
Molecular weight: 88.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A265QAA2|A0A265QAA2_9FIRM Oligopeptide ABC transporter ATP-binding protein OppF OS=Tissierella sp. P1 OX=1280483 GN=CIW83_03920 PE=3 SV=1
MM1 pKa = 7.69KK2 pKa = 10.23KK3 pKa = 10.44NKK5 pKa = 9.64IKK7 pKa = 10.59ILGLSIVLLVAMAGCRR23 pKa = 11.84PANTNMRR30 pKa = 11.84NMSTQTRR37 pKa = 11.84LYY39 pKa = 10.84NDD41 pKa = 3.42NLNNRR46 pKa = 11.84WMTNTPYY53 pKa = 9.63NTRR56 pKa = 11.84DD57 pKa = 3.44RR58 pKa = 11.84LNTNLNNGMVRR69 pKa = 11.84DD70 pKa = 3.98NTNLNNNALRR80 pKa = 11.84NTPMTYY86 pKa = 10.75DD87 pKa = 3.32SLRR90 pKa = 11.84GNNLVPGTTDD100 pKa = 2.95NGNIANNNLSTRR112 pKa = 11.84ATAIAKK118 pKa = 9.59RR119 pKa = 11.84VAEE122 pKa = 4.05LPEE125 pKa = 3.99VKK127 pKa = 9.96GASVIVHH134 pKa = 6.68GNTALWDD141 pKa = 3.82VNN143 pKa = 3.67
MM1 pKa = 7.69KK2 pKa = 10.23KK3 pKa = 10.44NKK5 pKa = 9.64IKK7 pKa = 10.59ILGLSIVLLVAMAGCRR23 pKa = 11.84PANTNMRR30 pKa = 11.84NMSTQTRR37 pKa = 11.84LYY39 pKa = 10.84NDD41 pKa = 3.42NLNNRR46 pKa = 11.84WMTNTPYY53 pKa = 9.63NTRR56 pKa = 11.84DD57 pKa = 3.44RR58 pKa = 11.84LNTNLNNGMVRR69 pKa = 11.84DD70 pKa = 3.98NTNLNNNALRR80 pKa = 11.84NTPMTYY86 pKa = 10.75DD87 pKa = 3.32SLRR90 pKa = 11.84GNNLVPGTTDD100 pKa = 2.95NGNIANNNLSTRR112 pKa = 11.84ATAIAKK118 pKa = 9.59RR119 pKa = 11.84VAEE122 pKa = 4.05LPEE125 pKa = 3.99VKK127 pKa = 9.96GASVIVHH134 pKa = 6.68GNTALWDD141 pKa = 3.82VNN143 pKa = 3.67
Molecular weight: 15.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1147079 |
20 |
1815 |
288.1 |
32.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.428 ± 0.045 | 0.904 ± 0.012 |
5.673 ± 0.032 | 7.777 ± 0.046 |
4.331 ± 0.028 | 6.604 ± 0.045 |
1.434 ± 0.013 | 10.508 ± 0.047 |
8.212 ± 0.038 | 9.333 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.737 ± 0.019 | 5.902 ± 0.03 |
2.984 ± 0.024 | 2.389 ± 0.019 |
3.696 ± 0.025 | 6.153 ± 0.028 |
4.916 ± 0.03 | 6.101 ± 0.034 |
0.756 ± 0.013 | 4.162 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
