
Ohlsdorf virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Ohlsrhavirus; Ohlsdorf ohlsrhavirus
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
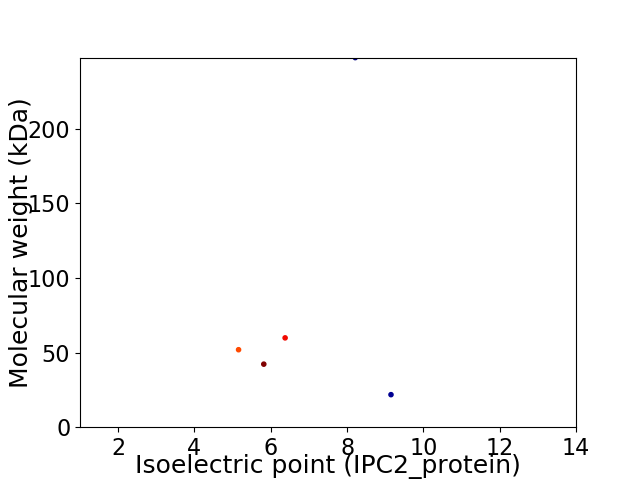
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A291I346|A0A291I346_9RHAB Nucleocapsid protein OS=Ohlsdorf virus OX=2040592 GN=N PE=4 SV=1
MM1 pKa = 7.69CIVDD5 pKa = 3.67QGCHH9 pKa = 4.76HH10 pKa = 7.15SIYY13 pKa = 10.55RR14 pKa = 11.84FLSTNLISVNMVVKK28 pKa = 10.52RR29 pKa = 11.84VSTDD33 pKa = 2.83PTKK36 pKa = 10.62RR37 pKa = 11.84VTVTHH42 pKa = 5.95MNIATDD48 pKa = 3.76RR49 pKa = 11.84PIAYY53 pKa = 9.14PSEE56 pKa = 4.15WFDD59 pKa = 4.18NPLNDD64 pKa = 4.47KK65 pKa = 10.69PKK67 pKa = 10.51ILIAAEE73 pKa = 3.99GDD75 pKa = 3.08ITRR78 pKa = 11.84AQLHH82 pKa = 5.85ATVQTNFPEE91 pKa = 4.81GDD93 pKa = 3.46LDD95 pKa = 3.64HH96 pKa = 6.95RR97 pKa = 11.84VAMMFLRR104 pKa = 11.84HH105 pKa = 4.56EE106 pKa = 4.49MMRR109 pKa = 11.84KK110 pKa = 8.48SHH112 pKa = 6.29MLGEE116 pKa = 4.7DD117 pKa = 2.78WTSFGRR123 pKa = 11.84AIGVRR128 pKa = 11.84GGDD131 pKa = 3.11ITIGNLVIFDD141 pKa = 3.82QTEE144 pKa = 3.9TRR146 pKa = 11.84PDD148 pKa = 3.59VIDD151 pKa = 5.64GEE153 pKa = 4.93DD154 pKa = 3.64CMTDD158 pKa = 3.51SEE160 pKa = 4.55VLKK163 pKa = 10.48YY164 pKa = 10.51IILICSMYY172 pKa = 10.67RR173 pKa = 11.84LAQISRR179 pKa = 11.84DD180 pKa = 3.63DD181 pKa = 3.5YY182 pKa = 10.44RR183 pKa = 11.84TNVVNNVTALIEE195 pKa = 4.41GLDD198 pKa = 3.81GEE200 pKa = 4.57AVEE203 pKa = 6.12FSTQILSYY211 pKa = 9.98RR212 pKa = 11.84KK213 pKa = 7.61WLAYY217 pKa = 9.86HH218 pKa = 7.12PYY220 pKa = 9.09IKK222 pKa = 10.23MMAAIDD228 pKa = 3.72MYY230 pKa = 11.28LNEE233 pKa = 4.66FPYY236 pKa = 10.19HH237 pKa = 5.93QYY239 pKa = 10.96ASARR243 pKa = 11.84IGTIVTRR250 pKa = 11.84FKK252 pKa = 11.02DD253 pKa = 3.62CSVLVATKK261 pKa = 9.93TITEE265 pKa = 4.07ALGLTFDD272 pKa = 5.25TFAEE276 pKa = 4.79WIWTARR282 pKa = 11.84CANQFVNVIKK292 pKa = 10.73GGEE295 pKa = 4.0EE296 pKa = 3.59MDD298 pKa = 3.66NQRR301 pKa = 11.84SYY303 pKa = 12.2SMYY306 pKa = 10.64FMDD309 pKa = 6.09FGLSPKK315 pKa = 10.01SPYY318 pKa = 10.13SATVNADD325 pKa = 2.96LHH327 pKa = 6.59LFYY330 pKa = 9.89HH331 pKa = 6.8TIGVSSGLEE340 pKa = 3.48RR341 pKa = 11.84SRR343 pKa = 11.84NARR346 pKa = 11.84YY347 pKa = 9.63IGNPEE352 pKa = 3.92HH353 pKa = 7.12NNIINNAMVIHH364 pKa = 5.84YY365 pKa = 10.03VLGTFSTFDD374 pKa = 3.33AQFSADD380 pKa = 3.5GVPIEE385 pKa = 5.38LEE387 pKa = 4.15ADD389 pKa = 3.72DD390 pKa = 6.15LVDD393 pKa = 6.19DD394 pKa = 5.57DD395 pKa = 6.02LPGDD399 pKa = 3.87NSPEE403 pKa = 3.65LWLGYY408 pKa = 9.94IISKK412 pKa = 9.99NGRR415 pKa = 11.84IPPIVARR422 pKa = 11.84QSILEE427 pKa = 4.09WNQFPGCRR435 pKa = 11.84EE436 pKa = 3.93SSIGLYY442 pKa = 9.92LYY444 pKa = 10.73KK445 pKa = 10.4RR446 pKa = 11.84AQTLMVSLVAPAQQ459 pKa = 3.42
MM1 pKa = 7.69CIVDD5 pKa = 3.67QGCHH9 pKa = 4.76HH10 pKa = 7.15SIYY13 pKa = 10.55RR14 pKa = 11.84FLSTNLISVNMVVKK28 pKa = 10.52RR29 pKa = 11.84VSTDD33 pKa = 2.83PTKK36 pKa = 10.62RR37 pKa = 11.84VTVTHH42 pKa = 5.95MNIATDD48 pKa = 3.76RR49 pKa = 11.84PIAYY53 pKa = 9.14PSEE56 pKa = 4.15WFDD59 pKa = 4.18NPLNDD64 pKa = 4.47KK65 pKa = 10.69PKK67 pKa = 10.51ILIAAEE73 pKa = 3.99GDD75 pKa = 3.08ITRR78 pKa = 11.84AQLHH82 pKa = 5.85ATVQTNFPEE91 pKa = 4.81GDD93 pKa = 3.46LDD95 pKa = 3.64HH96 pKa = 6.95RR97 pKa = 11.84VAMMFLRR104 pKa = 11.84HH105 pKa = 4.56EE106 pKa = 4.49MMRR109 pKa = 11.84KK110 pKa = 8.48SHH112 pKa = 6.29MLGEE116 pKa = 4.7DD117 pKa = 2.78WTSFGRR123 pKa = 11.84AIGVRR128 pKa = 11.84GGDD131 pKa = 3.11ITIGNLVIFDD141 pKa = 3.82QTEE144 pKa = 3.9TRR146 pKa = 11.84PDD148 pKa = 3.59VIDD151 pKa = 5.64GEE153 pKa = 4.93DD154 pKa = 3.64CMTDD158 pKa = 3.51SEE160 pKa = 4.55VLKK163 pKa = 10.48YY164 pKa = 10.51IILICSMYY172 pKa = 10.67RR173 pKa = 11.84LAQISRR179 pKa = 11.84DD180 pKa = 3.63DD181 pKa = 3.5YY182 pKa = 10.44RR183 pKa = 11.84TNVVNNVTALIEE195 pKa = 4.41GLDD198 pKa = 3.81GEE200 pKa = 4.57AVEE203 pKa = 6.12FSTQILSYY211 pKa = 9.98RR212 pKa = 11.84KK213 pKa = 7.61WLAYY217 pKa = 9.86HH218 pKa = 7.12PYY220 pKa = 9.09IKK222 pKa = 10.23MMAAIDD228 pKa = 3.72MYY230 pKa = 11.28LNEE233 pKa = 4.66FPYY236 pKa = 10.19HH237 pKa = 5.93QYY239 pKa = 10.96ASARR243 pKa = 11.84IGTIVTRR250 pKa = 11.84FKK252 pKa = 11.02DD253 pKa = 3.62CSVLVATKK261 pKa = 9.93TITEE265 pKa = 4.07ALGLTFDD272 pKa = 5.25TFAEE276 pKa = 4.79WIWTARR282 pKa = 11.84CANQFVNVIKK292 pKa = 10.73GGEE295 pKa = 4.0EE296 pKa = 3.59MDD298 pKa = 3.66NQRR301 pKa = 11.84SYY303 pKa = 12.2SMYY306 pKa = 10.64FMDD309 pKa = 6.09FGLSPKK315 pKa = 10.01SPYY318 pKa = 10.13SATVNADD325 pKa = 2.96LHH327 pKa = 6.59LFYY330 pKa = 9.89HH331 pKa = 6.8TIGVSSGLEE340 pKa = 3.48RR341 pKa = 11.84SRR343 pKa = 11.84NARR346 pKa = 11.84YY347 pKa = 9.63IGNPEE352 pKa = 3.92HH353 pKa = 7.12NNIINNAMVIHH364 pKa = 5.84YY365 pKa = 10.03VLGTFSTFDD374 pKa = 3.33AQFSADD380 pKa = 3.5GVPIEE385 pKa = 5.38LEE387 pKa = 4.15ADD389 pKa = 3.72DD390 pKa = 6.15LVDD393 pKa = 6.19DD394 pKa = 5.57DD395 pKa = 6.02LPGDD399 pKa = 3.87NSPEE403 pKa = 3.65LWLGYY408 pKa = 9.94IISKK412 pKa = 9.99NGRR415 pKa = 11.84IPPIVARR422 pKa = 11.84QSILEE427 pKa = 4.09WNQFPGCRR435 pKa = 11.84EE436 pKa = 3.93SSIGLYY442 pKa = 9.92LYY444 pKa = 10.73KK445 pKa = 10.4RR446 pKa = 11.84AQTLMVSLVAPAQQ459 pKa = 3.42
Molecular weight: 51.93 kDa
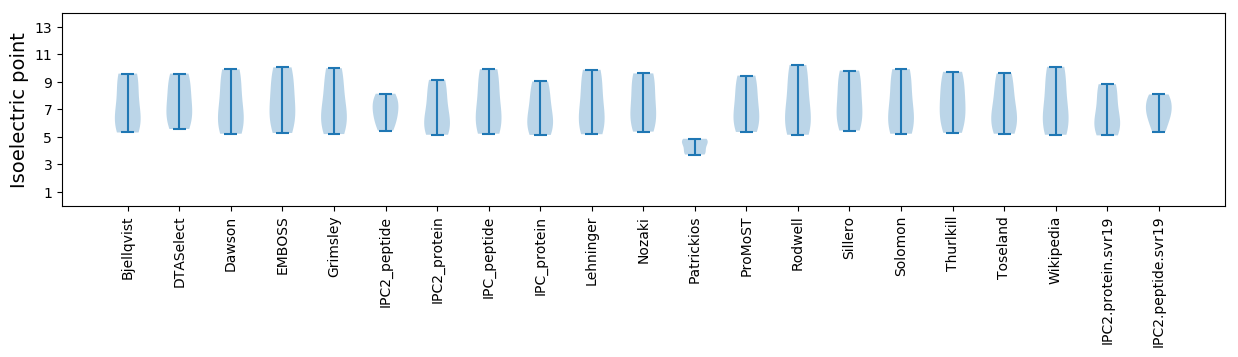
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A291I327|A0A291I327_9RHAB GDP polyribonucleotidyltransferase OS=Ohlsdorf virus OX=2040592 GN=L PE=4 SV=1
MM1 pKa = 7.49SSNSALLVAYY11 pKa = 9.05NLRR14 pKa = 11.84VNASLDD20 pKa = 3.35VKK22 pKa = 10.54FRR24 pKa = 11.84HH25 pKa = 6.26PPKK28 pKa = 10.28AYY30 pKa = 9.33KK31 pKa = 10.48DD32 pKa = 3.49LMRR35 pKa = 11.84ALEE38 pKa = 4.7LIKK41 pKa = 10.76DD42 pKa = 3.69NYY44 pKa = 11.03KK45 pKa = 10.87GFTRR49 pKa = 11.84DD50 pKa = 2.91KK51 pKa = 9.74EE52 pKa = 4.54LIFFFYY58 pKa = 10.94VMSGFFSKK66 pKa = 10.9YY67 pKa = 8.59INKK70 pKa = 9.71IGNFYY75 pKa = 10.69NYY77 pKa = 9.75QGTLSGVFNLMCKK90 pKa = 10.37NITPRR95 pKa = 11.84FPINYY100 pKa = 7.68EE101 pKa = 3.91FNGDD105 pKa = 3.97DD106 pKa = 5.08GGIHH110 pKa = 6.92PNFLITFHH118 pKa = 6.92LSVRR122 pKa = 11.84EE123 pKa = 3.75SDD125 pKa = 3.26MSYY128 pKa = 11.0QSLYY132 pKa = 10.98DD133 pKa = 3.48YY134 pKa = 10.13MKK136 pKa = 9.25EE137 pKa = 3.96HH138 pKa = 6.79SPSKK142 pKa = 8.92YY143 pKa = 9.47RR144 pKa = 11.84KK145 pKa = 7.97RR146 pKa = 11.84NKK148 pKa = 9.74VKK150 pKa = 10.16MSIFDD155 pKa = 4.61DD156 pKa = 4.33YY157 pKa = 11.18PVSITVKK164 pKa = 9.63QKK166 pKa = 10.5VFILTSIKK174 pKa = 10.67SSATKK179 pKa = 10.6NSFSPEE185 pKa = 3.65LKK187 pKa = 10.61
MM1 pKa = 7.49SSNSALLVAYY11 pKa = 9.05NLRR14 pKa = 11.84VNASLDD20 pKa = 3.35VKK22 pKa = 10.54FRR24 pKa = 11.84HH25 pKa = 6.26PPKK28 pKa = 10.28AYY30 pKa = 9.33KK31 pKa = 10.48DD32 pKa = 3.49LMRR35 pKa = 11.84ALEE38 pKa = 4.7LIKK41 pKa = 10.76DD42 pKa = 3.69NYY44 pKa = 11.03KK45 pKa = 10.87GFTRR49 pKa = 11.84DD50 pKa = 2.91KK51 pKa = 9.74EE52 pKa = 4.54LIFFFYY58 pKa = 10.94VMSGFFSKK66 pKa = 10.9YY67 pKa = 8.59INKK70 pKa = 9.71IGNFYY75 pKa = 10.69NYY77 pKa = 9.75QGTLSGVFNLMCKK90 pKa = 10.37NITPRR95 pKa = 11.84FPINYY100 pKa = 7.68EE101 pKa = 3.91FNGDD105 pKa = 3.97DD106 pKa = 5.08GGIHH110 pKa = 6.92PNFLITFHH118 pKa = 6.92LSVRR122 pKa = 11.84EE123 pKa = 3.75SDD125 pKa = 3.26MSYY128 pKa = 11.0QSLYY132 pKa = 10.98DD133 pKa = 3.48YY134 pKa = 10.13MKK136 pKa = 9.25EE137 pKa = 3.96HH138 pKa = 6.79SPSKK142 pKa = 8.92YY143 pKa = 9.47RR144 pKa = 11.84KK145 pKa = 7.97RR146 pKa = 11.84NKK148 pKa = 9.74VKK150 pKa = 10.16MSIFDD155 pKa = 4.61DD156 pKa = 4.33YY157 pKa = 11.18PVSITVKK164 pKa = 9.63QKK166 pKa = 10.5VFILTSIKK174 pKa = 10.67SSATKK179 pKa = 10.6NSFSPEE185 pKa = 3.65LKK187 pKa = 10.61
Molecular weight: 21.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3708 |
187 |
2160 |
741.6 |
84.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.585 ± 0.859 | 1.375 ± 0.257 |
5.556 ± 0.393 | 5.367 ± 0.424 |
4.396 ± 0.493 | 4.962 ± 0.375 |
2.778 ± 0.23 | 7.551 ± 0.621 |
6.823 ± 0.758 | 9.628 ± 1.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.697 ± 0.388 | 5.663 ± 0.544 |
4.746 ± 0.453 | 3.182 ± 0.371 |
4.773 ± 0.318 | 8.522 ± 0.445 |
6.095 ± 0.553 | 5.663 ± 0.444 |
1.591 ± 0.332 | 4.045 ± 0.45 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
