
Murine hepatitis virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Embecovirus; Murine coronavirus
Average proteome isoelectric point is 7.14
Get precalculated fractions of proteins
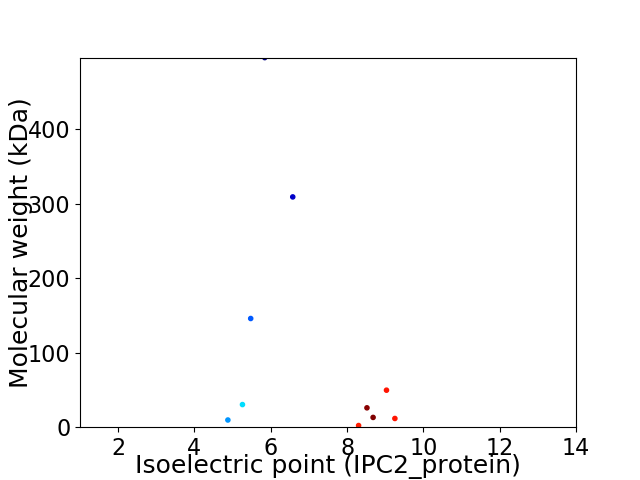
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q77NL4|Q77NL4_9BETC Non-structural protein 2a OS=Murine hepatitis virus OX=11138 GN=ORF2A PE=3 SV=1
MM1 pKa = 7.46FNLFLTDD8 pKa = 3.48TVWYY12 pKa = 8.66VGQIIFIFAVCLMVTIIVVAFLASIKK38 pKa = 10.74LCIQLCGLCNTLVLSPSIYY57 pKa = 10.4LYY59 pKa = 10.85DD60 pKa = 4.56RR61 pKa = 11.84SKK63 pKa = 11.14QLYY66 pKa = 8.86KK67 pKa = 10.82YY68 pKa = 9.71YY69 pKa = 10.26NEE71 pKa = 3.95EE72 pKa = 3.47MRR74 pKa = 11.84LPLLEE79 pKa = 4.79VDD81 pKa = 5.76DD82 pKa = 4.29II83 pKa = 4.66
MM1 pKa = 7.46FNLFLTDD8 pKa = 3.48TVWYY12 pKa = 8.66VGQIIFIFAVCLMVTIIVVAFLASIKK38 pKa = 10.74LCIQLCGLCNTLVLSPSIYY57 pKa = 10.4LYY59 pKa = 10.85DD60 pKa = 4.56RR61 pKa = 11.84SKK63 pKa = 11.14QLYY66 pKa = 8.86KK67 pKa = 10.82YY68 pKa = 9.71YY69 pKa = 10.26NEE71 pKa = 3.95EE72 pKa = 3.47MRR74 pKa = 11.84LPLLEE79 pKa = 4.79VDD81 pKa = 5.76DD82 pKa = 4.29II83 pKa = 4.66
Molecular weight: 9.67 kDa
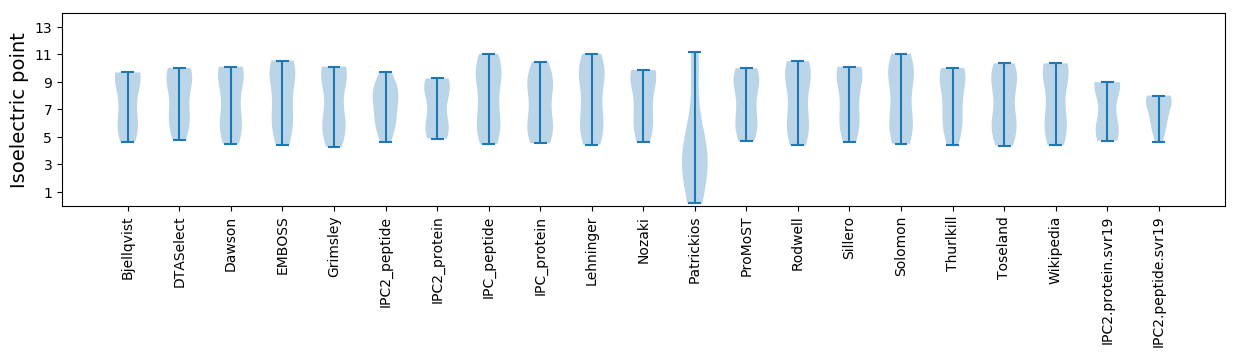
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9J3E3|Q9J3E3_9BETC Membrane protein OS=Murine hepatitis virus OX=11138 GN=ORF6 PE=3 SV=1
MM1 pKa = 7.35SFVPGQEE8 pKa = 3.97NAGGRR13 pKa = 11.84SSSVNRR19 pKa = 11.84AGNGILKK26 pKa = 8.59KK27 pKa = 7.37TTWADD32 pKa = 3.24QTEE35 pKa = 4.28RR36 pKa = 11.84GPNNQNRR43 pKa = 11.84GRR45 pKa = 11.84RR46 pKa = 11.84NQPKK50 pKa = 8.18QTATTQPNSGSVVPHH65 pKa = 5.66YY66 pKa = 11.05SWFSGITQFQKK77 pKa = 10.82GKK79 pKa = 9.62EE80 pKa = 3.93FQFAEE85 pKa = 4.59GQGVPIANGIPASEE99 pKa = 4.21QKK101 pKa = 10.87GYY103 pKa = 9.69WYY105 pKa = 10.34RR106 pKa = 11.84HH107 pKa = 4.36NRR109 pKa = 11.84RR110 pKa = 11.84SFKK113 pKa = 10.75TPDD116 pKa = 3.34GQQKK120 pKa = 9.76QLLPRR125 pKa = 11.84WYY127 pKa = 9.79FYY129 pKa = 11.56YY130 pKa = 10.78LGTGPHH136 pKa = 6.72AGASYY141 pKa = 10.53GDD143 pKa = 4.18SIEE146 pKa = 4.14GVFWVANSQADD157 pKa = 3.73TNTRR161 pKa = 11.84SDD163 pKa = 3.07IVEE166 pKa = 4.41RR167 pKa = 11.84DD168 pKa = 3.66PSSHH172 pKa = 5.75EE173 pKa = 4.53AIPTRR178 pKa = 11.84FAPGTVLPQGFYY190 pKa = 10.92VEE192 pKa = 4.42GSGRR196 pKa = 11.84SAPASRR202 pKa = 11.84SGSRR206 pKa = 11.84SQSRR210 pKa = 11.84GPNNRR215 pKa = 11.84ARR217 pKa = 11.84SSSNQRR223 pKa = 11.84QPASTVKK230 pKa = 10.23PDD232 pKa = 3.37MAEE235 pKa = 4.03EE236 pKa = 3.7IAALVLAKK244 pKa = 10.33LGKK247 pKa = 10.0DD248 pKa = 2.88AGQPKK253 pKa = 9.61QVTKK257 pKa = 10.71QSAKK261 pKa = 8.91EE262 pKa = 3.89VRR264 pKa = 11.84QKK266 pKa = 10.73ILNKK270 pKa = 9.17PRR272 pKa = 11.84QKK274 pKa = 9.65RR275 pKa = 11.84TPNKK279 pKa = 8.97QCPVQQCFGKK289 pKa = 10.52RR290 pKa = 11.84GPNQNFGGSEE300 pKa = 3.94MLKK303 pKa = 10.6LGTSDD308 pKa = 3.38PQFPILAEE316 pKa = 3.97LAPTVGAFFFGSKK329 pKa = 10.69LEE331 pKa = 4.03LVKK334 pKa = 10.9KK335 pKa = 10.59NSGGADD341 pKa = 3.41EE342 pKa = 4.48PTKK345 pKa = 10.65DD346 pKa = 3.71VYY348 pKa = 10.84EE349 pKa = 4.14LQYY352 pKa = 11.13SGAVRR357 pKa = 11.84FDD359 pKa = 3.21STLPGFEE366 pKa = 4.83TIMKK370 pKa = 9.51VLNEE374 pKa = 4.26NLNAYY379 pKa = 9.5QKK381 pKa = 11.1DD382 pKa = 3.25GGADD386 pKa = 3.39VVSPKK391 pKa = 9.52PQRR394 pKa = 11.84KK395 pKa = 8.65GRR397 pKa = 11.84RR398 pKa = 11.84QALEE402 pKa = 4.0KK403 pKa = 10.3KK404 pKa = 10.6DD405 pKa = 3.86EE406 pKa = 4.05VDD408 pKa = 3.31NVSVAKK414 pKa = 10.14PKK416 pKa = 10.81SSVQRR421 pKa = 11.84NVSRR425 pKa = 11.84EE426 pKa = 4.02LTPEE430 pKa = 4.02DD431 pKa = 4.12RR432 pKa = 11.84SLLAQILDD440 pKa = 4.15DD441 pKa = 4.33GVVPDD446 pKa = 4.88GLEE449 pKa = 4.2DD450 pKa = 4.13DD451 pKa = 4.85SNVV454 pKa = 3.06
MM1 pKa = 7.35SFVPGQEE8 pKa = 3.97NAGGRR13 pKa = 11.84SSSVNRR19 pKa = 11.84AGNGILKK26 pKa = 8.59KK27 pKa = 7.37TTWADD32 pKa = 3.24QTEE35 pKa = 4.28RR36 pKa = 11.84GPNNQNRR43 pKa = 11.84GRR45 pKa = 11.84RR46 pKa = 11.84NQPKK50 pKa = 8.18QTATTQPNSGSVVPHH65 pKa = 5.66YY66 pKa = 11.05SWFSGITQFQKK77 pKa = 10.82GKK79 pKa = 9.62EE80 pKa = 3.93FQFAEE85 pKa = 4.59GQGVPIANGIPASEE99 pKa = 4.21QKK101 pKa = 10.87GYY103 pKa = 9.69WYY105 pKa = 10.34RR106 pKa = 11.84HH107 pKa = 4.36NRR109 pKa = 11.84RR110 pKa = 11.84SFKK113 pKa = 10.75TPDD116 pKa = 3.34GQQKK120 pKa = 9.76QLLPRR125 pKa = 11.84WYY127 pKa = 9.79FYY129 pKa = 11.56YY130 pKa = 10.78LGTGPHH136 pKa = 6.72AGASYY141 pKa = 10.53GDD143 pKa = 4.18SIEE146 pKa = 4.14GVFWVANSQADD157 pKa = 3.73TNTRR161 pKa = 11.84SDD163 pKa = 3.07IVEE166 pKa = 4.41RR167 pKa = 11.84DD168 pKa = 3.66PSSHH172 pKa = 5.75EE173 pKa = 4.53AIPTRR178 pKa = 11.84FAPGTVLPQGFYY190 pKa = 10.92VEE192 pKa = 4.42GSGRR196 pKa = 11.84SAPASRR202 pKa = 11.84SGSRR206 pKa = 11.84SQSRR210 pKa = 11.84GPNNRR215 pKa = 11.84ARR217 pKa = 11.84SSSNQRR223 pKa = 11.84QPASTVKK230 pKa = 10.23PDD232 pKa = 3.37MAEE235 pKa = 4.03EE236 pKa = 3.7IAALVLAKK244 pKa = 10.33LGKK247 pKa = 10.0DD248 pKa = 2.88AGQPKK253 pKa = 9.61QVTKK257 pKa = 10.71QSAKK261 pKa = 8.91EE262 pKa = 3.89VRR264 pKa = 11.84QKK266 pKa = 10.73ILNKK270 pKa = 9.17PRR272 pKa = 11.84QKK274 pKa = 9.65RR275 pKa = 11.84TPNKK279 pKa = 8.97QCPVQQCFGKK289 pKa = 10.52RR290 pKa = 11.84GPNQNFGGSEE300 pKa = 3.94MLKK303 pKa = 10.6LGTSDD308 pKa = 3.38PQFPILAEE316 pKa = 3.97LAPTVGAFFFGSKK329 pKa = 10.69LEE331 pKa = 4.03LVKK334 pKa = 10.9KK335 pKa = 10.59NSGGADD341 pKa = 3.41EE342 pKa = 4.48PTKK345 pKa = 10.65DD346 pKa = 3.71VYY348 pKa = 10.84EE349 pKa = 4.14LQYY352 pKa = 11.13SGAVRR357 pKa = 11.84FDD359 pKa = 3.21STLPGFEE366 pKa = 4.83TIMKK370 pKa = 9.51VLNEE374 pKa = 4.26NLNAYY379 pKa = 9.5QKK381 pKa = 11.1DD382 pKa = 3.25GGADD386 pKa = 3.39VVSPKK391 pKa = 9.52PQRR394 pKa = 11.84KK395 pKa = 8.65GRR397 pKa = 11.84RR398 pKa = 11.84QALEE402 pKa = 4.0KK403 pKa = 10.3KK404 pKa = 10.6DD405 pKa = 3.86EE406 pKa = 4.05VDD408 pKa = 3.31NVSVAKK414 pKa = 10.14PKK416 pKa = 10.81SSVQRR421 pKa = 11.84NVSRR425 pKa = 11.84EE426 pKa = 4.02LTPEE430 pKa = 4.02DD431 pKa = 4.12RR432 pKa = 11.84SLLAQILDD440 pKa = 4.15DD441 pKa = 4.33GVVPDD446 pKa = 4.88GLEE449 pKa = 4.2DD450 pKa = 4.13DD451 pKa = 4.85SNVV454 pKa = 3.06
Molecular weight: 49.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9787 |
19 |
4467 |
978.7 |
109.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.805 ± 0.355 | 3.75 ± 0.329 |
5.589 ± 0.28 | 4.23 ± 0.186 |
5.344 ± 0.142 | 6.151 ± 0.293 |
1.778 ± 0.187 | 4.455 ± 0.493 |
5.712 ± 0.361 | 9.094 ± 0.251 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.156 ± 0.233 | 5.099 ± 0.611 |
3.832 ± 0.323 | 3.505 ± 0.374 |
3.852 ± 0.402 | 7.101 ± 0.266 |
5.783 ± 0.267 | 9.666 ± 0.93 |
1.318 ± 0.117 | 4.782 ± 0.313 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
