
Chickpea chlorotic stunt virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
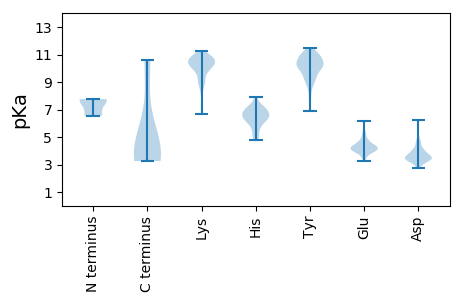
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|Q17S72|Q17S72_9LUTE p0 protein OS=Chickpea chlorotic stunt virus OX=328430 GN=p0 PE=4 SV=1
MM1 pKa = 7.29NVLINKK7 pKa = 6.81FTSNFEE13 pKa = 4.54INFSPSLSLATRR25 pKa = 11.84RR26 pKa = 11.84SLICLFLANSSEE38 pKa = 4.32FIKK41 pKa = 10.61ISNEE45 pKa = 3.39RR46 pKa = 11.84DD47 pKa = 3.39GTDD50 pKa = 2.73KK51 pKa = 10.4STCYY55 pKa = 10.13RR56 pKa = 11.84SLLLLLPFLFGSFEE70 pKa = 4.07FFGDD74 pKa = 3.54QLVVPWNGGNNRR86 pKa = 11.84HH87 pKa = 6.39HH88 pKa = 7.13GLFQRR93 pKa = 11.84LLTNTRR99 pKa = 11.84FCTLRR104 pKa = 11.84LQPGRR109 pKa = 11.84EE110 pKa = 3.74ARR112 pKa = 11.84YY113 pKa = 9.77NPSIQITPTRR123 pKa = 11.84VEE125 pKa = 4.35LYY127 pKa = 10.21RR128 pKa = 11.84YY129 pKa = 8.4IQSPLVEE136 pKa = 4.51GLSGHH141 pKa = 6.32QDD143 pKa = 3.49LLSLGLHH150 pKa = 6.61HH151 pKa = 6.43FTKK154 pKa = 10.87LVGAYY159 pKa = 9.19VRR161 pKa = 11.84QLEE164 pKa = 4.34RR165 pKa = 11.84NNFCLRR171 pKa = 11.84SEE173 pKa = 4.7IILGDD178 pKa = 3.95SVDD181 pKa = 4.61LDD183 pKa = 3.94VSHH186 pKa = 7.36LGILLMDD193 pKa = 4.84GSDD196 pKa = 4.14HH197 pKa = 7.02NNTHH201 pKa = 6.47HH202 pKa = 7.17AYY204 pKa = 10.19SGSRR208 pKa = 11.84VINCLHH214 pKa = 6.5RR215 pKa = 11.84CYY217 pKa = 10.72GQALTSDD224 pKa = 2.86IWEE227 pKa = 4.39LFGVDD232 pKa = 4.03YY233 pKa = 10.37CPCLQVPSFPLEE245 pKa = 3.82AEE247 pKa = 3.89IPEE250 pKa = 4.94DD251 pKa = 3.53SFQFDD256 pKa = 3.72EE257 pKa = 5.04DD258 pKa = 4.42SEE260 pKa = 5.18GEE262 pKa = 3.92DD263 pKa = 5.63DD264 pKa = 4.05EE265 pKa = 6.16GFWEE269 pKa = 4.36LL270 pKa = 4.69
MM1 pKa = 7.29NVLINKK7 pKa = 6.81FTSNFEE13 pKa = 4.54INFSPSLSLATRR25 pKa = 11.84RR26 pKa = 11.84SLICLFLANSSEE38 pKa = 4.32FIKK41 pKa = 10.61ISNEE45 pKa = 3.39RR46 pKa = 11.84DD47 pKa = 3.39GTDD50 pKa = 2.73KK51 pKa = 10.4STCYY55 pKa = 10.13RR56 pKa = 11.84SLLLLLPFLFGSFEE70 pKa = 4.07FFGDD74 pKa = 3.54QLVVPWNGGNNRR86 pKa = 11.84HH87 pKa = 6.39HH88 pKa = 7.13GLFQRR93 pKa = 11.84LLTNTRR99 pKa = 11.84FCTLRR104 pKa = 11.84LQPGRR109 pKa = 11.84EE110 pKa = 3.74ARR112 pKa = 11.84YY113 pKa = 9.77NPSIQITPTRR123 pKa = 11.84VEE125 pKa = 4.35LYY127 pKa = 10.21RR128 pKa = 11.84YY129 pKa = 8.4IQSPLVEE136 pKa = 4.51GLSGHH141 pKa = 6.32QDD143 pKa = 3.49LLSLGLHH150 pKa = 6.61HH151 pKa = 6.43FTKK154 pKa = 10.87LVGAYY159 pKa = 9.19VRR161 pKa = 11.84QLEE164 pKa = 4.34RR165 pKa = 11.84NNFCLRR171 pKa = 11.84SEE173 pKa = 4.7IILGDD178 pKa = 3.95SVDD181 pKa = 4.61LDD183 pKa = 3.94VSHH186 pKa = 7.36LGILLMDD193 pKa = 4.84GSDD196 pKa = 4.14HH197 pKa = 7.02NNTHH201 pKa = 6.47HH202 pKa = 7.17AYY204 pKa = 10.19SGSRR208 pKa = 11.84VINCLHH214 pKa = 6.5RR215 pKa = 11.84CYY217 pKa = 10.72GQALTSDD224 pKa = 2.86IWEE227 pKa = 4.39LFGVDD232 pKa = 4.03YY233 pKa = 10.37CPCLQVPSFPLEE245 pKa = 3.82AEE247 pKa = 3.89IPEE250 pKa = 4.94DD251 pKa = 3.53SFQFDD256 pKa = 3.72EE257 pKa = 5.04DD258 pKa = 4.42SEE260 pKa = 5.18GEE262 pKa = 3.92DD263 pKa = 5.63DD264 pKa = 4.05EE265 pKa = 6.16GFWEE269 pKa = 4.36LL270 pKa = 4.69
Molecular weight: 30.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q17S69|Q17S69_9LUTE Readthrough protein OS=Chickpea chlorotic stunt virus OX=328430 GN=p5 PE=3 SV=1
MM1 pKa = 6.53NTVVVRR7 pKa = 11.84NNGRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84NRR16 pKa = 11.84RR17 pKa = 11.84TVQRR21 pKa = 11.84ARR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84NPVVVVEE32 pKa = 5.36APRR35 pKa = 11.84QPQRR39 pKa = 11.84GRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84NRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84ASGRR53 pKa = 11.84STAGRR58 pKa = 11.84RR59 pKa = 11.84GSSEE63 pKa = 3.82TFVFSKK69 pKa = 11.13DD70 pKa = 3.3NLAGSSSGSITFGPSLSDD88 pKa = 3.49CPAFSSGILRR98 pKa = 11.84AYY100 pKa = 10.24HH101 pKa = 6.47EE102 pKa = 4.67YY103 pKa = 9.99KK104 pKa = 10.23ISMVKK109 pKa = 10.66LEE111 pKa = 5.26FISEE115 pKa = 4.18AASTSSGSIAYY126 pKa = 9.43EE127 pKa = 3.86LDD129 pKa = 3.43PHH131 pKa = 6.83CKK133 pKa = 8.5STSLGSYY140 pKa = 9.12INKK143 pKa = 9.91FGITSNGQRR152 pKa = 11.84TFAARR157 pKa = 11.84LINGIEE163 pKa = 3.93WHH165 pKa = 6.9SSDD168 pKa = 3.83EE169 pKa = 4.08DD170 pKa = 3.46QFRR173 pKa = 11.84ILYY176 pKa = 9.29KK177 pKa = 10.97GNGGSAIAGSFRR189 pKa = 11.84ITIKK193 pKa = 10.84CQTQNPKK200 pKa = 10.57
MM1 pKa = 6.53NTVVVRR7 pKa = 11.84NNGRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84NRR16 pKa = 11.84RR17 pKa = 11.84TVQRR21 pKa = 11.84ARR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84NPVVVVEE32 pKa = 5.36APRR35 pKa = 11.84QPQRR39 pKa = 11.84GRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84NRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84ASGRR53 pKa = 11.84STAGRR58 pKa = 11.84RR59 pKa = 11.84GSSEE63 pKa = 3.82TFVFSKK69 pKa = 11.13DD70 pKa = 3.3NLAGSSSGSITFGPSLSDD88 pKa = 3.49CPAFSSGILRR98 pKa = 11.84AYY100 pKa = 10.24HH101 pKa = 6.47EE102 pKa = 4.67YY103 pKa = 9.99KK104 pKa = 10.23ISMVKK109 pKa = 10.66LEE111 pKa = 5.26FISEE115 pKa = 4.18AASTSSGSIAYY126 pKa = 9.43EE127 pKa = 3.86LDD129 pKa = 3.43PHH131 pKa = 6.83CKK133 pKa = 8.5STSLGSYY140 pKa = 9.12INKK143 pKa = 9.91FGITSNGQRR152 pKa = 11.84TFAARR157 pKa = 11.84LINGIEE163 pKa = 3.93WHH165 pKa = 6.9SSDD168 pKa = 3.83EE169 pKa = 4.08DD170 pKa = 3.46QFRR173 pKa = 11.84ILYY176 pKa = 9.29KK177 pKa = 10.97GNGGSAIAGSFRR189 pKa = 11.84ITIKK193 pKa = 10.84CQTQNPKK200 pKa = 10.57
Molecular weight: 22.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3109 |
193 |
1093 |
518.2 |
58.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.143 ± 0.448 | 1.866 ± 0.249 |
4.439 ± 0.418 | 6.819 ± 0.515 |
4.085 ± 0.414 | 7.044 ± 0.393 |
1.994 ± 0.286 | 4.728 ± 0.251 |
6.143 ± 0.848 | 8.684 ± 1.093 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.898 ± 0.263 | 4.76 ± 0.377 |
5.05 ± 0.457 | 3.763 ± 0.206 |
6.304 ± 1.119 | 9.682 ± 0.71 |
5.436 ± 0.462 | 5.886 ± 0.449 |
1.994 ± 0.254 | 3.249 ± 0.249 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
