
Dickeya phage BF25/12
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Corkvirinae; Stompvirus; Dickeya virus BF25-12
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
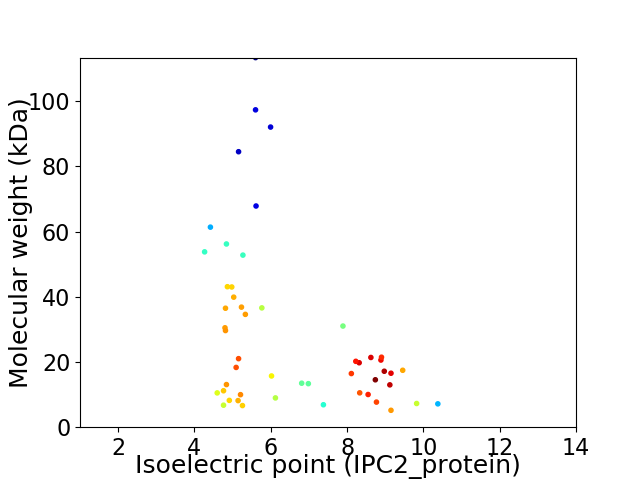
Virtual 2D-PAGE plot for 51 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A219MH13|A0A219MH13_9CAUD Putative DNA exonuclease OS=Dickeya phage BF25/12 OX=1698708 GN=BF2512_30 PE=4 SV=1
MM1 pKa = 7.88KK2 pKa = 9.23ITIAEE7 pKa = 4.14PVLINPLEE15 pKa = 4.12QFKK18 pKa = 9.01PTVATNTRR26 pKa = 11.84KK27 pKa = 9.93LLAEE31 pKa = 4.15IFLARR36 pKa = 11.84DD37 pKa = 3.52TTTVEE42 pKa = 3.87AQPLDD47 pKa = 3.8KK48 pKa = 10.9EE49 pKa = 4.56VLKK52 pKa = 10.78EE53 pKa = 3.95MKK55 pKa = 10.06QLVYY59 pKa = 9.99TGCDD63 pKa = 2.75NKK65 pKa = 10.2MVSQYY70 pKa = 10.87SEE72 pKa = 4.29HH73 pKa = 7.28PSPEE77 pKa = 3.52AHH79 pKa = 6.68MLRR82 pKa = 11.84AVGVAASVADD92 pKa = 3.95FEE94 pKa = 5.08EE95 pKa = 4.25NRR97 pKa = 11.84EE98 pKa = 3.47GSYY101 pKa = 9.95RR102 pKa = 11.84VEE104 pKa = 5.77FAFQWLRR111 pKa = 11.84GYY113 pKa = 10.35LFKK116 pKa = 11.43NNLPMPTEE124 pKa = 4.66FPANTLGALSYY135 pKa = 9.75PARR138 pKa = 11.84TCCCMDD144 pKa = 5.04DD145 pKa = 4.15YY146 pKa = 11.03LSSMRR151 pKa = 11.84VIPSKK156 pKa = 10.78LLYY159 pKa = 9.45HH160 pKa = 6.87LKK162 pKa = 10.33EE163 pKa = 4.08AVPPVLLAMYY173 pKa = 8.66EE174 pKa = 4.15PRR176 pKa = 11.84TQSYY180 pKa = 9.04YY181 pKa = 11.33GKK183 pKa = 10.26DD184 pKa = 2.98LVEE187 pKa = 4.63RR188 pKa = 11.84QTEE191 pKa = 4.13VNNIGGYY198 pKa = 9.79CGVVYY203 pKa = 10.53TEE205 pKa = 6.08DD206 pKa = 3.21GTAWFAGFLNADD218 pKa = 3.94DD219 pKa = 3.87VLKK222 pKa = 11.09LRR224 pKa = 11.84FRR226 pKa = 11.84RR227 pKa = 11.84MRR229 pKa = 11.84LGAAIKK235 pKa = 10.33EE236 pKa = 4.17LTGDD240 pKa = 3.72DD241 pKa = 3.53NLARR245 pKa = 11.84KK246 pKa = 8.86AADD249 pKa = 3.09IRR251 pKa = 11.84DD252 pKa = 3.63LSSYY256 pKa = 11.48DD257 pKa = 4.4LVLHH261 pKa = 6.5PNDD264 pKa = 3.9RR265 pKa = 11.84PFGDD269 pKa = 3.4EE270 pKa = 3.86YY271 pKa = 11.41VRR273 pKa = 11.84MRR275 pKa = 11.84EE276 pKa = 3.97EE277 pKa = 4.49GNSLEE282 pKa = 4.03SCMAHH287 pKa = 6.42SAGNYY292 pKa = 7.28DD293 pKa = 3.56APFNVHH299 pKa = 6.32PCDD302 pKa = 3.89VYY304 pKa = 10.17SCAYY308 pKa = 9.62YY309 pKa = 10.55GQGDD313 pKa = 3.68NGLVLVEE320 pKa = 4.23AQQGGIPVGRR330 pKa = 11.84GILNVHH336 pKa = 6.29TKK338 pKa = 10.52QIVRR342 pKa = 11.84WYY344 pKa = 9.71GEE346 pKa = 3.93YY347 pKa = 10.12KK348 pKa = 10.59ASVMLKK354 pKa = 7.91NTFGIKK360 pKa = 9.92VDD362 pKa = 4.04GDD364 pKa = 3.86ALDD367 pKa = 4.81DD368 pKa = 3.83VQLALIQDD376 pKa = 4.12GAKK379 pKa = 9.78IAAPYY384 pKa = 10.71LDD386 pKa = 4.48GCQACDD392 pKa = 3.32IEE394 pKa = 6.07DD395 pKa = 3.82GEE397 pKa = 4.82LYY399 pKa = 10.36VRR401 pKa = 11.84HH402 pKa = 6.47RR403 pKa = 11.84GDD405 pKa = 2.9IVMDD409 pKa = 3.83DD410 pKa = 3.18TSGYY414 pKa = 8.7QDD416 pKa = 3.24CKK418 pKa = 10.97ARR420 pKa = 11.84EE421 pKa = 4.22LDD423 pKa = 3.87TLSGDD428 pKa = 4.1YY429 pKa = 10.82YY430 pKa = 10.47PEE432 pKa = 5.21SDD434 pKa = 5.27LEE436 pKa = 4.07YY437 pKa = 10.91QPIHH441 pKa = 6.05DD442 pKa = 4.27TYY444 pKa = 10.97YY445 pKa = 11.17NPDD448 pKa = 3.78NTHH451 pKa = 6.05MGYY454 pKa = 10.23FCPITEE460 pKa = 4.7EE461 pKa = 4.26YY462 pKa = 9.65CNPYY466 pKa = 8.45ATSAAYY472 pKa = 10.48VDD474 pKa = 4.51GEE476 pKa = 4.36VTRR479 pKa = 11.84ISDD482 pKa = 3.49VVYY485 pKa = 10.02FGYY488 pKa = 10.37INPPGSWEE496 pKa = 3.94NLGGSIGWTEE506 pKa = 3.6NSEE509 pKa = 4.27GYY511 pKa = 10.5SYY513 pKa = 11.55DD514 pKa = 3.93DD515 pKa = 4.06EE516 pKa = 4.56TEE518 pKa = 3.38EE519 pKa = 4.61WYY521 pKa = 9.7TNEE524 pKa = 5.4QYY526 pKa = 11.14DD527 pKa = 3.47QLIAEE532 pKa = 4.46RR533 pKa = 11.84EE534 pKa = 4.05EE535 pKa = 3.98QEE537 pKa = 3.91EE538 pKa = 4.46DD539 pKa = 3.31KK540 pKa = 11.71DD541 pKa = 3.78EE542 pKa = 4.28AAA544 pKa = 5.0
MM1 pKa = 7.88KK2 pKa = 9.23ITIAEE7 pKa = 4.14PVLINPLEE15 pKa = 4.12QFKK18 pKa = 9.01PTVATNTRR26 pKa = 11.84KK27 pKa = 9.93LLAEE31 pKa = 4.15IFLARR36 pKa = 11.84DD37 pKa = 3.52TTTVEE42 pKa = 3.87AQPLDD47 pKa = 3.8KK48 pKa = 10.9EE49 pKa = 4.56VLKK52 pKa = 10.78EE53 pKa = 3.95MKK55 pKa = 10.06QLVYY59 pKa = 9.99TGCDD63 pKa = 2.75NKK65 pKa = 10.2MVSQYY70 pKa = 10.87SEE72 pKa = 4.29HH73 pKa = 7.28PSPEE77 pKa = 3.52AHH79 pKa = 6.68MLRR82 pKa = 11.84AVGVAASVADD92 pKa = 3.95FEE94 pKa = 5.08EE95 pKa = 4.25NRR97 pKa = 11.84EE98 pKa = 3.47GSYY101 pKa = 9.95RR102 pKa = 11.84VEE104 pKa = 5.77FAFQWLRR111 pKa = 11.84GYY113 pKa = 10.35LFKK116 pKa = 11.43NNLPMPTEE124 pKa = 4.66FPANTLGALSYY135 pKa = 9.75PARR138 pKa = 11.84TCCCMDD144 pKa = 5.04DD145 pKa = 4.15YY146 pKa = 11.03LSSMRR151 pKa = 11.84VIPSKK156 pKa = 10.78LLYY159 pKa = 9.45HH160 pKa = 6.87LKK162 pKa = 10.33EE163 pKa = 4.08AVPPVLLAMYY173 pKa = 8.66EE174 pKa = 4.15PRR176 pKa = 11.84TQSYY180 pKa = 9.04YY181 pKa = 11.33GKK183 pKa = 10.26DD184 pKa = 2.98LVEE187 pKa = 4.63RR188 pKa = 11.84QTEE191 pKa = 4.13VNNIGGYY198 pKa = 9.79CGVVYY203 pKa = 10.53TEE205 pKa = 6.08DD206 pKa = 3.21GTAWFAGFLNADD218 pKa = 3.94DD219 pKa = 3.87VLKK222 pKa = 11.09LRR224 pKa = 11.84FRR226 pKa = 11.84RR227 pKa = 11.84MRR229 pKa = 11.84LGAAIKK235 pKa = 10.33EE236 pKa = 4.17LTGDD240 pKa = 3.72DD241 pKa = 3.53NLARR245 pKa = 11.84KK246 pKa = 8.86AADD249 pKa = 3.09IRR251 pKa = 11.84DD252 pKa = 3.63LSSYY256 pKa = 11.48DD257 pKa = 4.4LVLHH261 pKa = 6.5PNDD264 pKa = 3.9RR265 pKa = 11.84PFGDD269 pKa = 3.4EE270 pKa = 3.86YY271 pKa = 11.41VRR273 pKa = 11.84MRR275 pKa = 11.84EE276 pKa = 3.97EE277 pKa = 4.49GNSLEE282 pKa = 4.03SCMAHH287 pKa = 6.42SAGNYY292 pKa = 7.28DD293 pKa = 3.56APFNVHH299 pKa = 6.32PCDD302 pKa = 3.89VYY304 pKa = 10.17SCAYY308 pKa = 9.62YY309 pKa = 10.55GQGDD313 pKa = 3.68NGLVLVEE320 pKa = 4.23AQQGGIPVGRR330 pKa = 11.84GILNVHH336 pKa = 6.29TKK338 pKa = 10.52QIVRR342 pKa = 11.84WYY344 pKa = 9.71GEE346 pKa = 3.93YY347 pKa = 10.12KK348 pKa = 10.59ASVMLKK354 pKa = 7.91NTFGIKK360 pKa = 9.92VDD362 pKa = 4.04GDD364 pKa = 3.86ALDD367 pKa = 4.81DD368 pKa = 3.83VQLALIQDD376 pKa = 4.12GAKK379 pKa = 9.78IAAPYY384 pKa = 10.71LDD386 pKa = 4.48GCQACDD392 pKa = 3.32IEE394 pKa = 6.07DD395 pKa = 3.82GEE397 pKa = 4.82LYY399 pKa = 10.36VRR401 pKa = 11.84HH402 pKa = 6.47RR403 pKa = 11.84GDD405 pKa = 2.9IVMDD409 pKa = 3.83DD410 pKa = 3.18TSGYY414 pKa = 8.7QDD416 pKa = 3.24CKK418 pKa = 10.97ARR420 pKa = 11.84EE421 pKa = 4.22LDD423 pKa = 3.87TLSGDD428 pKa = 4.1YY429 pKa = 10.82YY430 pKa = 10.47PEE432 pKa = 5.21SDD434 pKa = 5.27LEE436 pKa = 4.07YY437 pKa = 10.91QPIHH441 pKa = 6.05DD442 pKa = 4.27TYY444 pKa = 10.97YY445 pKa = 11.17NPDD448 pKa = 3.78NTHH451 pKa = 6.05MGYY454 pKa = 10.23FCPITEE460 pKa = 4.7EE461 pKa = 4.26YY462 pKa = 9.65CNPYY466 pKa = 8.45ATSAAYY472 pKa = 10.48VDD474 pKa = 4.51GEE476 pKa = 4.36VTRR479 pKa = 11.84ISDD482 pKa = 3.49VVYY485 pKa = 10.02FGYY488 pKa = 10.37INPPGSWEE496 pKa = 3.94NLGGSIGWTEE506 pKa = 3.6NSEE509 pKa = 4.27GYY511 pKa = 10.5SYY513 pKa = 11.55DD514 pKa = 3.93DD515 pKa = 4.06EE516 pKa = 4.56TEE518 pKa = 3.38EE519 pKa = 4.61WYY521 pKa = 9.7TNEE524 pKa = 5.4QYY526 pKa = 11.14DD527 pKa = 3.47QLIAEE532 pKa = 4.46RR533 pKa = 11.84EE534 pKa = 4.05EE535 pKa = 3.98QEE537 pKa = 3.91EE538 pKa = 4.46DD539 pKa = 3.31KK540 pKa = 11.71DD541 pKa = 3.78EE542 pKa = 4.28AAA544 pKa = 5.0
Molecular weight: 61.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A219MH35|A0A219MH35_9CAUD Uncharacterized protein OS=Dickeya phage BF25/12 OX=1698708 GN=BF2512_51 PE=4 SV=1
MM1 pKa = 7.74KK2 pKa = 10.0DD3 pKa = 3.49YY4 pKa = 11.73LEE6 pKa = 4.5VCSTSRR12 pKa = 11.84TGLRR16 pKa = 11.84WKK18 pKa = 9.98QRR20 pKa = 11.84PKK22 pKa = 10.46QSSVTPGEE30 pKa = 3.93PAFTSVNGKK39 pKa = 9.98GYY41 pKa = 10.5YY42 pKa = 9.56EE43 pKa = 5.15GYY45 pKa = 10.48FKK47 pKa = 11.03GKK49 pKa = 9.67RR50 pKa = 11.84YY51 pKa = 9.05LAHH54 pKa = 6.79RR55 pKa = 11.84VIVYY59 pKa = 9.6LLSGKK64 pKa = 9.05WPEE67 pKa = 4.22QVDD70 pKa = 4.22HH71 pKa = 7.29IDD73 pKa = 3.64GNRR76 pKa = 11.84SNNCPTNLRR85 pKa = 11.84AVTAAEE91 pKa = 3.96NQHH94 pKa = 6.04NKK96 pKa = 9.24LANGFKK102 pKa = 10.39LDD104 pKa = 3.82PRR106 pKa = 11.84VGRR109 pKa = 11.84YY110 pKa = 6.38QARR113 pKa = 11.84IRR115 pKa = 11.84VDD117 pKa = 3.45GKK119 pKa = 10.16QVNLGSFATKK129 pKa = 10.35QEE131 pKa = 3.71AQAAYY136 pKa = 9.74LAAKK140 pKa = 8.36RR141 pKa = 11.84TYY143 pKa = 10.51HH144 pKa = 5.38PTAPEE149 pKa = 3.68RR150 pKa = 11.84CYY152 pKa = 11.43VKK154 pKa = 10.86
MM1 pKa = 7.74KK2 pKa = 10.0DD3 pKa = 3.49YY4 pKa = 11.73LEE6 pKa = 4.5VCSTSRR12 pKa = 11.84TGLRR16 pKa = 11.84WKK18 pKa = 9.98QRR20 pKa = 11.84PKK22 pKa = 10.46QSSVTPGEE30 pKa = 3.93PAFTSVNGKK39 pKa = 9.98GYY41 pKa = 10.5YY42 pKa = 9.56EE43 pKa = 5.15GYY45 pKa = 10.48FKK47 pKa = 11.03GKK49 pKa = 9.67RR50 pKa = 11.84YY51 pKa = 9.05LAHH54 pKa = 6.79RR55 pKa = 11.84VIVYY59 pKa = 9.6LLSGKK64 pKa = 9.05WPEE67 pKa = 4.22QVDD70 pKa = 4.22HH71 pKa = 7.29IDD73 pKa = 3.64GNRR76 pKa = 11.84SNNCPTNLRR85 pKa = 11.84AVTAAEE91 pKa = 3.96NQHH94 pKa = 6.04NKK96 pKa = 9.24LANGFKK102 pKa = 10.39LDD104 pKa = 3.82PRR106 pKa = 11.84VGRR109 pKa = 11.84YY110 pKa = 6.38QARR113 pKa = 11.84IRR115 pKa = 11.84VDD117 pKa = 3.45GKK119 pKa = 10.16QVNLGSFATKK129 pKa = 10.35QEE131 pKa = 3.71AQAAYY136 pKa = 9.74LAAKK140 pKa = 8.36RR141 pKa = 11.84TYY143 pKa = 10.51HH144 pKa = 5.38PTAPEE149 pKa = 3.68RR150 pKa = 11.84CYY152 pKa = 11.43VKK154 pKa = 10.86
Molecular weight: 17.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13224 |
46 |
1039 |
259.3 |
28.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.725 ± 0.434 | 1.195 ± 0.177 |
6.322 ± 0.236 | 5.203 ± 0.277 |
3.176 ± 0.172 | 7.698 ± 0.252 |
2.178 ± 0.225 | 4.545 ± 0.189 |
4.515 ± 0.296 | 8.719 ± 0.217 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.715 ± 0.178 | 4.583 ± 0.225 |
4.114 ± 0.198 | 4.537 ± 0.353 |
5.74 ± 0.262 | 6.178 ± 0.258 |
6.496 ± 0.439 | 7.35 ± 0.2 |
1.338 ± 0.112 | 3.675 ± 0.244 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
