
Lewinella aurantiaca
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Saprospiria; Saprospirales; Lewinellaceae; Lewinella
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
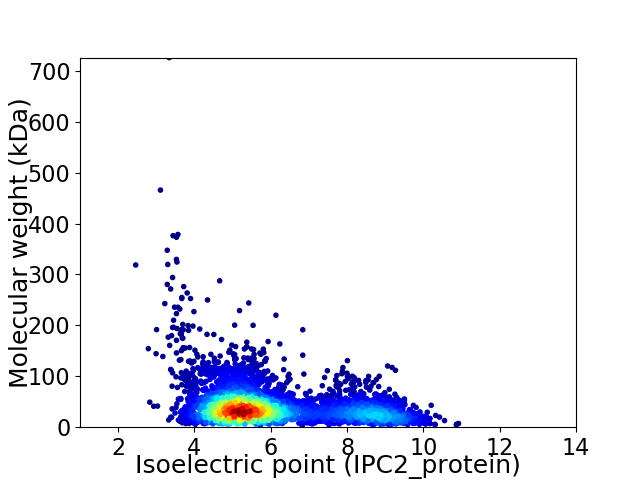
Virtual 2D-PAGE plot for 4341 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C7G0F6|A0A5C7G0F6_9BACT Ligase-associated DNA damage response exonuclease OS=Lewinella aurantiaca OX=2602767 GN=FUA23_02695 PE=4 SV=1
MM1 pKa = 7.21YY2 pKa = 10.31KK3 pKa = 10.33VNCLLLSTMLMLTLANASLLAQGTTYY29 pKa = 10.5EE30 pKa = 4.28VNFASIAFVNLEE42 pKa = 4.24LNEE45 pKa = 4.46DD46 pKa = 3.78CTRR49 pKa = 11.84DD50 pKa = 3.77VIYY53 pKa = 10.73MNLLTGNSDD62 pKa = 2.89VDD64 pKa = 3.49GDD66 pKa = 4.64GIVPPQEE73 pKa = 3.69AFIIEE78 pKa = 4.22IDD80 pKa = 4.88DD81 pKa = 3.94NSPDD85 pKa = 3.48NGPIVDD91 pKa = 3.95GCGMFNYY98 pKa = 9.53TIRR101 pKa = 11.84PNPDD105 pKa = 2.66SSVVGFTFGSGTITARR121 pKa = 11.84DD122 pKa = 3.81ATPPQQFGTATPNIGPFFTPQLQEE146 pKa = 3.83LTINTLPASISRR158 pKa = 11.84TFMVDD163 pKa = 3.03GASGFPLMNSLSGEE177 pKa = 4.01LMNRR181 pKa = 11.84LLAGGIIPRR190 pKa = 11.84FTDD193 pKa = 2.93ACSDD197 pKa = 3.7VTVVVSDD204 pKa = 4.27EE205 pKa = 4.45IINTGACEE213 pKa = 4.22DD214 pKa = 3.76IIIRR218 pKa = 11.84RR219 pKa = 11.84SFMATDD225 pKa = 4.1ASADD229 pKa = 3.99CISPNPAGNNGTTTVTYY246 pKa = 10.17DD247 pKa = 3.15IVLEE251 pKa = 4.37RR252 pKa = 11.84PNSANVIAPIDD263 pKa = 3.83LVTYY267 pKa = 8.86EE268 pKa = 4.8CNDD271 pKa = 3.53PALAGGNFPDD281 pKa = 5.49PDD283 pKa = 4.69PEE285 pKa = 5.52DD286 pKa = 3.85YY287 pKa = 11.0PFLNGPNGPVYY298 pKa = 10.88LNEE301 pKa = 3.86IYY303 pKa = 11.09GNVGASFTNSGAVQICNNTIKK324 pKa = 10.63YY325 pKa = 8.82VRR327 pKa = 11.84TYY329 pKa = 10.4TVIDD333 pKa = 3.28WCDD336 pKa = 3.11TDD338 pKa = 4.09NVQTFTQLVKK348 pKa = 11.04VGDD351 pKa = 3.86TGAPTIVAPTQDD363 pKa = 4.31LDD365 pKa = 4.39FDD367 pKa = 5.08LLPDD371 pKa = 4.66DD372 pKa = 4.9GPLVFSTNAPGCGAYY387 pKa = 8.78LTTNMAGLSITDD399 pKa = 3.56GCSALMTVTAFVLINGDD416 pKa = 3.47EE417 pKa = 4.22DD418 pKa = 4.39NISGAINVYY427 pKa = 10.44ASSATDD433 pKa = 3.07RR434 pKa = 11.84LTPFLPAGPHH444 pKa = 6.33IIRR447 pKa = 11.84YY448 pKa = 7.68VAEE451 pKa = 4.6DD452 pKa = 3.31EE453 pKa = 5.09CGNQTTSDD461 pKa = 3.59LDD463 pKa = 3.42IVVEE467 pKa = 4.21DD468 pKa = 3.71RR469 pKa = 11.84SGPVMIVEE477 pKa = 4.55DD478 pKa = 4.37ALNVALSSSGFATVNATDD496 pKa = 4.59LDD498 pKa = 4.0EE499 pKa = 5.85GSYY502 pKa = 11.3DD503 pKa = 3.74DD504 pKa = 4.68CTDD507 pKa = 2.82ITLEE511 pKa = 3.93IAFANINSLMPIGAYY526 pKa = 10.02GPSITLSCVDD536 pKa = 4.56LDD538 pKa = 3.92QVVTGIPVIIRR549 pKa = 11.84GTDD552 pKa = 3.1EE553 pKa = 4.18NGNTNTRR560 pKa = 11.84MSVLNVVDD568 pKa = 3.92NTAPICIAPGNLNLTCSEE586 pKa = 4.22ADD588 pKa = 3.02AMLPEE593 pKa = 5.05DD594 pKa = 3.93VNAVYY599 pKa = 8.54NTDD602 pKa = 3.5PEE604 pKa = 4.69GTVQMFDD611 pKa = 3.22QLFGEE616 pKa = 4.69VTTLDD621 pKa = 3.46NCGNEE626 pKa = 3.83YY627 pKa = 10.45SAQNIVANINDD638 pKa = 4.71CGTGTINRR646 pKa = 11.84SFTVTDD652 pKa = 3.75GQGFVSAPGCEE663 pKa = 4.01QIISIRR669 pKa = 11.84GVRR672 pKa = 11.84NYY674 pKa = 9.02TVEE677 pKa = 4.27FPGDD681 pKa = 3.31ASATCGLMPDD691 pKa = 3.81VNDD694 pKa = 4.46FVYY697 pKa = 10.55TPLGCDD703 pKa = 2.93MVVSYY708 pKa = 11.35VEE710 pKa = 3.71VDD712 pKa = 3.47TFFAASDD719 pKa = 3.42ACFKK723 pKa = 10.33IRR725 pKa = 11.84RR726 pKa = 11.84TIEE729 pKa = 3.8IINWCEE735 pKa = 3.29YY736 pKa = 10.89DD737 pKa = 3.77GNGDD741 pKa = 4.07FYY743 pKa = 10.5TIRR746 pKa = 11.84RR747 pKa = 11.84DD748 pKa = 3.17ADD750 pKa = 3.14NDD752 pKa = 3.67GNYY755 pKa = 10.29EE756 pKa = 3.98EE757 pKa = 4.97STFLHH762 pKa = 6.61VIPNGNTDD770 pKa = 3.31AGDD773 pKa = 3.79DD774 pKa = 4.1VAVLDD779 pKa = 4.67QDD781 pKa = 4.14SDD783 pKa = 3.7RR784 pKa = 11.84DD785 pKa = 3.7NLNNIGFLDD794 pKa = 3.53VDD796 pKa = 4.41DD797 pKa = 5.01NFGAFGTDD805 pKa = 2.89SDD807 pKa = 4.29NDD809 pKa = 3.73GDD811 pKa = 3.73TGYY814 pKa = 11.37ADD816 pKa = 3.56SEE818 pKa = 4.46SRR820 pKa = 11.84GAFRR824 pKa = 11.84YY825 pKa = 8.7VQFIKK830 pKa = 10.8VYY832 pKa = 10.97DD833 pKa = 4.44NIAPTITNISSDD845 pKa = 3.54VANSEE850 pKa = 4.16DD851 pKa = 4.28CNGGGIQIDD860 pKa = 3.81YY861 pKa = 9.02TITDD865 pKa = 3.75DD866 pKa = 4.76CVGANLSTNVEE877 pKa = 3.83LDD879 pKa = 3.61LNFVAGGGFSATRR892 pKa = 11.84EE893 pKa = 4.16LTSDD897 pKa = 3.56EE898 pKa = 4.63VIGDD902 pKa = 3.49GNGNYY907 pKa = 10.03NVILSGLPVGEE918 pKa = 3.91HH919 pKa = 7.52AIRR922 pKa = 11.84VAATDD927 pKa = 3.22GCGNVNGRR935 pKa = 11.84IIQFDD940 pKa = 3.4IQDD943 pKa = 3.86NSTVTPICIGRR954 pKa = 11.84LTFVLMNDD962 pKa = 3.68GAGGGSALVEE972 pKa = 4.01ADD974 pKa = 3.96DD975 pKa = 5.45FVVSLNGNCNNVDD988 pKa = 3.01VDD990 pKa = 3.46YY991 pKa = 11.31SIYY994 pKa = 10.41RR995 pKa = 11.84DD996 pKa = 3.78FEE998 pKa = 4.75YY999 pKa = 10.94NDD1001 pKa = 3.78PNFVPGTNRR1010 pKa = 11.84TDD1012 pKa = 3.95FPVSCDD1018 pKa = 3.46DD1019 pKa = 3.65VGDD1022 pKa = 4.04ILVQVYY1028 pKa = 10.34AFTPNGQAGLCNATAVIEE1046 pKa = 4.13PSSTVSCTAANVASLSGFITSPRR1069 pKa = 11.84NEE1071 pKa = 4.31LLDD1074 pKa = 4.64GIEE1077 pKa = 3.99VHH1079 pKa = 6.53ISDD1082 pKa = 5.01MDD1084 pKa = 3.76TMDD1087 pKa = 4.82DD1088 pKa = 4.1MLYY1091 pKa = 10.06TDD1093 pKa = 4.71ANGSFLFPALSEE1105 pKa = 3.81GHH1107 pKa = 6.56EE1108 pKa = 4.09YY1109 pKa = 9.92MIRR1112 pKa = 11.84PSMPDD1117 pKa = 3.03EE1118 pKa = 4.5VNLEE1122 pKa = 4.23RR1123 pKa = 11.84IKK1125 pKa = 10.37TSDD1128 pKa = 2.69ITIIGAHH1135 pKa = 6.49ALGAILIEE1143 pKa = 4.53DD1144 pKa = 5.2PYY1146 pKa = 11.88RR1147 pKa = 11.84MIAADD1152 pKa = 3.61ANGDD1156 pKa = 3.73GFIDD1160 pKa = 3.8IGDD1163 pKa = 4.15MIAIRR1168 pKa = 11.84RR1169 pKa = 11.84VILGIDD1175 pKa = 3.29ATFTEE1180 pKa = 5.32GPTWRR1185 pKa = 11.84FIRR1188 pKa = 11.84RR1189 pKa = 11.84DD1190 pKa = 3.29FDD1192 pKa = 4.94LEE1194 pKa = 4.04GLTEE1198 pKa = 4.28GWDD1201 pKa = 3.3PDD1203 pKa = 4.14VFPTTYY1209 pKa = 10.0RR1210 pKa = 11.84VEE1212 pKa = 4.05EE1213 pKa = 4.01LLGHH1217 pKa = 5.75NRR1219 pKa = 11.84EE1220 pKa = 3.98ADD1222 pKa = 3.87FVAIEE1227 pKa = 4.06IGDD1230 pKa = 3.86VFVQSTGRR1238 pKa = 11.84EE1239 pKa = 4.28SQPLTASDD1247 pKa = 3.09ATLAVGEE1254 pKa = 5.02RR1255 pKa = 11.84IDD1257 pKa = 5.11LDD1259 pKa = 3.17ITARR1263 pKa = 11.84DD1264 pKa = 3.29LAGFQGTFDD1273 pKa = 3.64AASGLEE1279 pKa = 3.84IEE1281 pKa = 4.81GWSSEE1286 pKa = 4.07VLSAGNVNDD1295 pKa = 4.43RR1296 pKa = 11.84YY1297 pKa = 10.65LSHH1300 pKa = 6.85GLLAVSYY1307 pKa = 10.77NGDD1310 pKa = 3.78EE1311 pKa = 4.78LEE1313 pKa = 4.9DD1314 pKa = 3.82NAPVLTLHH1322 pKa = 6.66LRR1324 pKa = 11.84AAAPDD1329 pKa = 3.75VRR1331 pKa = 11.84ISDD1334 pKa = 4.05YY1335 pKa = 11.51LSVTDD1340 pKa = 4.18RR1341 pKa = 11.84MTYY1344 pKa = 10.19PEE1346 pKa = 5.45AILTGGNTATLSLEE1360 pKa = 4.26FSEE1363 pKa = 4.82TTAGGADD1370 pKa = 3.81VILHH1374 pKa = 5.56QNFPNPVAAQTNIVFEE1390 pKa = 4.57LPRR1393 pKa = 11.84ASDD1396 pKa = 3.5VKK1398 pKa = 11.34LEE1400 pKa = 3.92VHH1402 pKa = 6.91DD1403 pKa = 3.99LQGRR1407 pKa = 11.84IVTKK1411 pKa = 10.59RR1412 pKa = 11.84SLEE1415 pKa = 4.09GQAGRR1420 pKa = 11.84NTITLSTFDD1429 pKa = 4.98DD1430 pKa = 4.15LNNYY1434 pKa = 8.42TGVLTYY1440 pKa = 10.03TITVGQTRR1448 pKa = 11.84LTKK1451 pKa = 10.67RR1452 pKa = 11.84MTVVAARR1459 pKa = 3.78
MM1 pKa = 7.21YY2 pKa = 10.31KK3 pKa = 10.33VNCLLLSTMLMLTLANASLLAQGTTYY29 pKa = 10.5EE30 pKa = 4.28VNFASIAFVNLEE42 pKa = 4.24LNEE45 pKa = 4.46DD46 pKa = 3.78CTRR49 pKa = 11.84DD50 pKa = 3.77VIYY53 pKa = 10.73MNLLTGNSDD62 pKa = 2.89VDD64 pKa = 3.49GDD66 pKa = 4.64GIVPPQEE73 pKa = 3.69AFIIEE78 pKa = 4.22IDD80 pKa = 4.88DD81 pKa = 3.94NSPDD85 pKa = 3.48NGPIVDD91 pKa = 3.95GCGMFNYY98 pKa = 9.53TIRR101 pKa = 11.84PNPDD105 pKa = 2.66SSVVGFTFGSGTITARR121 pKa = 11.84DD122 pKa = 3.81ATPPQQFGTATPNIGPFFTPQLQEE146 pKa = 3.83LTINTLPASISRR158 pKa = 11.84TFMVDD163 pKa = 3.03GASGFPLMNSLSGEE177 pKa = 4.01LMNRR181 pKa = 11.84LLAGGIIPRR190 pKa = 11.84FTDD193 pKa = 2.93ACSDD197 pKa = 3.7VTVVVSDD204 pKa = 4.27EE205 pKa = 4.45IINTGACEE213 pKa = 4.22DD214 pKa = 3.76IIIRR218 pKa = 11.84RR219 pKa = 11.84SFMATDD225 pKa = 4.1ASADD229 pKa = 3.99CISPNPAGNNGTTTVTYY246 pKa = 10.17DD247 pKa = 3.15IVLEE251 pKa = 4.37RR252 pKa = 11.84PNSANVIAPIDD263 pKa = 3.83LVTYY267 pKa = 8.86EE268 pKa = 4.8CNDD271 pKa = 3.53PALAGGNFPDD281 pKa = 5.49PDD283 pKa = 4.69PEE285 pKa = 5.52DD286 pKa = 3.85YY287 pKa = 11.0PFLNGPNGPVYY298 pKa = 10.88LNEE301 pKa = 3.86IYY303 pKa = 11.09GNVGASFTNSGAVQICNNTIKK324 pKa = 10.63YY325 pKa = 8.82VRR327 pKa = 11.84TYY329 pKa = 10.4TVIDD333 pKa = 3.28WCDD336 pKa = 3.11TDD338 pKa = 4.09NVQTFTQLVKK348 pKa = 11.04VGDD351 pKa = 3.86TGAPTIVAPTQDD363 pKa = 4.31LDD365 pKa = 4.39FDD367 pKa = 5.08LLPDD371 pKa = 4.66DD372 pKa = 4.9GPLVFSTNAPGCGAYY387 pKa = 8.78LTTNMAGLSITDD399 pKa = 3.56GCSALMTVTAFVLINGDD416 pKa = 3.47EE417 pKa = 4.22DD418 pKa = 4.39NISGAINVYY427 pKa = 10.44ASSATDD433 pKa = 3.07RR434 pKa = 11.84LTPFLPAGPHH444 pKa = 6.33IIRR447 pKa = 11.84YY448 pKa = 7.68VAEE451 pKa = 4.6DD452 pKa = 3.31EE453 pKa = 5.09CGNQTTSDD461 pKa = 3.59LDD463 pKa = 3.42IVVEE467 pKa = 4.21DD468 pKa = 3.71RR469 pKa = 11.84SGPVMIVEE477 pKa = 4.55DD478 pKa = 4.37ALNVALSSSGFATVNATDD496 pKa = 4.59LDD498 pKa = 4.0EE499 pKa = 5.85GSYY502 pKa = 11.3DD503 pKa = 3.74DD504 pKa = 4.68CTDD507 pKa = 2.82ITLEE511 pKa = 3.93IAFANINSLMPIGAYY526 pKa = 10.02GPSITLSCVDD536 pKa = 4.56LDD538 pKa = 3.92QVVTGIPVIIRR549 pKa = 11.84GTDD552 pKa = 3.1EE553 pKa = 4.18NGNTNTRR560 pKa = 11.84MSVLNVVDD568 pKa = 3.92NTAPICIAPGNLNLTCSEE586 pKa = 4.22ADD588 pKa = 3.02AMLPEE593 pKa = 5.05DD594 pKa = 3.93VNAVYY599 pKa = 8.54NTDD602 pKa = 3.5PEE604 pKa = 4.69GTVQMFDD611 pKa = 3.22QLFGEE616 pKa = 4.69VTTLDD621 pKa = 3.46NCGNEE626 pKa = 3.83YY627 pKa = 10.45SAQNIVANINDD638 pKa = 4.71CGTGTINRR646 pKa = 11.84SFTVTDD652 pKa = 3.75GQGFVSAPGCEE663 pKa = 4.01QIISIRR669 pKa = 11.84GVRR672 pKa = 11.84NYY674 pKa = 9.02TVEE677 pKa = 4.27FPGDD681 pKa = 3.31ASATCGLMPDD691 pKa = 3.81VNDD694 pKa = 4.46FVYY697 pKa = 10.55TPLGCDD703 pKa = 2.93MVVSYY708 pKa = 11.35VEE710 pKa = 3.71VDD712 pKa = 3.47TFFAASDD719 pKa = 3.42ACFKK723 pKa = 10.33IRR725 pKa = 11.84RR726 pKa = 11.84TIEE729 pKa = 3.8IINWCEE735 pKa = 3.29YY736 pKa = 10.89DD737 pKa = 3.77GNGDD741 pKa = 4.07FYY743 pKa = 10.5TIRR746 pKa = 11.84RR747 pKa = 11.84DD748 pKa = 3.17ADD750 pKa = 3.14NDD752 pKa = 3.67GNYY755 pKa = 10.29EE756 pKa = 3.98EE757 pKa = 4.97STFLHH762 pKa = 6.61VIPNGNTDD770 pKa = 3.31AGDD773 pKa = 3.79DD774 pKa = 4.1VAVLDD779 pKa = 4.67QDD781 pKa = 4.14SDD783 pKa = 3.7RR784 pKa = 11.84DD785 pKa = 3.7NLNNIGFLDD794 pKa = 3.53VDD796 pKa = 4.41DD797 pKa = 5.01NFGAFGTDD805 pKa = 2.89SDD807 pKa = 4.29NDD809 pKa = 3.73GDD811 pKa = 3.73TGYY814 pKa = 11.37ADD816 pKa = 3.56SEE818 pKa = 4.46SRR820 pKa = 11.84GAFRR824 pKa = 11.84YY825 pKa = 8.7VQFIKK830 pKa = 10.8VYY832 pKa = 10.97DD833 pKa = 4.44NIAPTITNISSDD845 pKa = 3.54VANSEE850 pKa = 4.16DD851 pKa = 4.28CNGGGIQIDD860 pKa = 3.81YY861 pKa = 9.02TITDD865 pKa = 3.75DD866 pKa = 4.76CVGANLSTNVEE877 pKa = 3.83LDD879 pKa = 3.61LNFVAGGGFSATRR892 pKa = 11.84EE893 pKa = 4.16LTSDD897 pKa = 3.56EE898 pKa = 4.63VIGDD902 pKa = 3.49GNGNYY907 pKa = 10.03NVILSGLPVGEE918 pKa = 3.91HH919 pKa = 7.52AIRR922 pKa = 11.84VAATDD927 pKa = 3.22GCGNVNGRR935 pKa = 11.84IIQFDD940 pKa = 3.4IQDD943 pKa = 3.86NSTVTPICIGRR954 pKa = 11.84LTFVLMNDD962 pKa = 3.68GAGGGSALVEE972 pKa = 4.01ADD974 pKa = 3.96DD975 pKa = 5.45FVVSLNGNCNNVDD988 pKa = 3.01VDD990 pKa = 3.46YY991 pKa = 11.31SIYY994 pKa = 10.41RR995 pKa = 11.84DD996 pKa = 3.78FEE998 pKa = 4.75YY999 pKa = 10.94NDD1001 pKa = 3.78PNFVPGTNRR1010 pKa = 11.84TDD1012 pKa = 3.95FPVSCDD1018 pKa = 3.46DD1019 pKa = 3.65VGDD1022 pKa = 4.04ILVQVYY1028 pKa = 10.34AFTPNGQAGLCNATAVIEE1046 pKa = 4.13PSSTVSCTAANVASLSGFITSPRR1069 pKa = 11.84NEE1071 pKa = 4.31LLDD1074 pKa = 4.64GIEE1077 pKa = 3.99VHH1079 pKa = 6.53ISDD1082 pKa = 5.01MDD1084 pKa = 3.76TMDD1087 pKa = 4.82DD1088 pKa = 4.1MLYY1091 pKa = 10.06TDD1093 pKa = 4.71ANGSFLFPALSEE1105 pKa = 3.81GHH1107 pKa = 6.56EE1108 pKa = 4.09YY1109 pKa = 9.92MIRR1112 pKa = 11.84PSMPDD1117 pKa = 3.03EE1118 pKa = 4.5VNLEE1122 pKa = 4.23RR1123 pKa = 11.84IKK1125 pKa = 10.37TSDD1128 pKa = 2.69ITIIGAHH1135 pKa = 6.49ALGAILIEE1143 pKa = 4.53DD1144 pKa = 5.2PYY1146 pKa = 11.88RR1147 pKa = 11.84MIAADD1152 pKa = 3.61ANGDD1156 pKa = 3.73GFIDD1160 pKa = 3.8IGDD1163 pKa = 4.15MIAIRR1168 pKa = 11.84RR1169 pKa = 11.84VILGIDD1175 pKa = 3.29ATFTEE1180 pKa = 5.32GPTWRR1185 pKa = 11.84FIRR1188 pKa = 11.84RR1189 pKa = 11.84DD1190 pKa = 3.29FDD1192 pKa = 4.94LEE1194 pKa = 4.04GLTEE1198 pKa = 4.28GWDD1201 pKa = 3.3PDD1203 pKa = 4.14VFPTTYY1209 pKa = 10.0RR1210 pKa = 11.84VEE1212 pKa = 4.05EE1213 pKa = 4.01LLGHH1217 pKa = 5.75NRR1219 pKa = 11.84EE1220 pKa = 3.98ADD1222 pKa = 3.87FVAIEE1227 pKa = 4.06IGDD1230 pKa = 3.86VFVQSTGRR1238 pKa = 11.84EE1239 pKa = 4.28SQPLTASDD1247 pKa = 3.09ATLAVGEE1254 pKa = 5.02RR1255 pKa = 11.84IDD1257 pKa = 5.11LDD1259 pKa = 3.17ITARR1263 pKa = 11.84DD1264 pKa = 3.29LAGFQGTFDD1273 pKa = 3.64AASGLEE1279 pKa = 3.84IEE1281 pKa = 4.81GWSSEE1286 pKa = 4.07VLSAGNVNDD1295 pKa = 4.43RR1296 pKa = 11.84YY1297 pKa = 10.65LSHH1300 pKa = 6.85GLLAVSYY1307 pKa = 10.77NGDD1310 pKa = 3.78EE1311 pKa = 4.78LEE1313 pKa = 4.9DD1314 pKa = 3.82NAPVLTLHH1322 pKa = 6.66LRR1324 pKa = 11.84AAAPDD1329 pKa = 3.75VRR1331 pKa = 11.84ISDD1334 pKa = 4.05YY1335 pKa = 11.51LSVTDD1340 pKa = 4.18RR1341 pKa = 11.84MTYY1344 pKa = 10.19PEE1346 pKa = 5.45AILTGGNTATLSLEE1360 pKa = 4.26FSEE1363 pKa = 4.82TTAGGADD1370 pKa = 3.81VILHH1374 pKa = 5.56QNFPNPVAAQTNIVFEE1390 pKa = 4.57LPRR1393 pKa = 11.84ASDD1396 pKa = 3.5VKK1398 pKa = 11.34LEE1400 pKa = 3.92VHH1402 pKa = 6.91DD1403 pKa = 3.99LQGRR1407 pKa = 11.84IVTKK1411 pKa = 10.59RR1412 pKa = 11.84SLEE1415 pKa = 4.09GQAGRR1420 pKa = 11.84NTITLSTFDD1429 pKa = 4.98DD1430 pKa = 4.15LNNYY1434 pKa = 8.42TGVLTYY1440 pKa = 10.03TITVGQTRR1448 pKa = 11.84LTKK1451 pKa = 10.67RR1452 pKa = 11.84MTVVAARR1459 pKa = 3.78
Molecular weight: 156.17 kDa
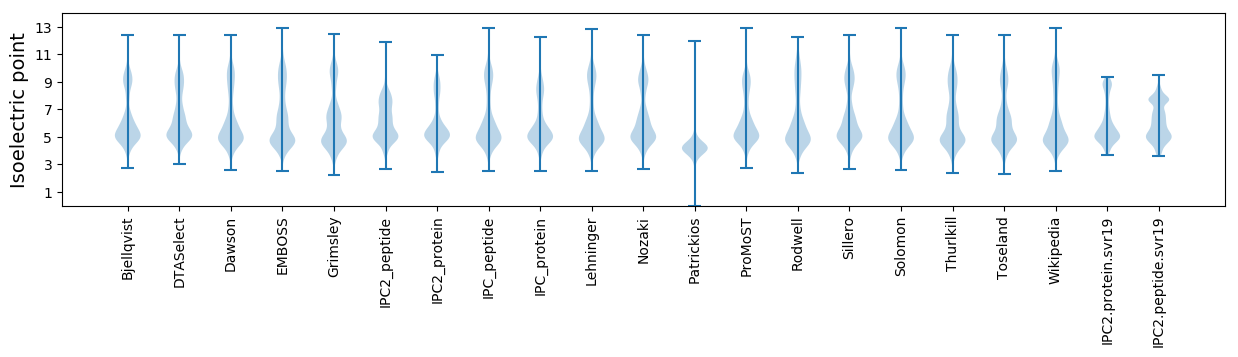
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C7F2H3|A0A5C7F2H3_9BACT DUF4221 domain-containing protein OS=Lewinella aurantiaca OX=2602767 GN=FUA23_21990 PE=4 SV=1
MM1 pKa = 6.98TNHH4 pKa = 5.74YY5 pKa = 7.38RR6 pKa = 11.84TCSRR10 pKa = 11.84LASGGQRR17 pKa = 11.84TASFPYY23 pKa = 9.84RR24 pKa = 11.84LAAVFFDD31 pKa = 3.5VSRR34 pKa = 11.84VSGVGLPTLLGICLLPLLLLCSGCGEE60 pKa = 4.5KK61 pKa = 10.79DD62 pKa = 3.06PAADD66 pKa = 4.52AEE68 pKa = 4.6VTTMKK73 pKa = 10.72ASAKK77 pKa = 9.08PEE79 pKa = 3.85PSAAEE84 pKa = 3.82IPLVTLHH91 pKa = 5.99TATRR95 pKa = 11.84GRR97 pKa = 11.84LPLRR101 pKa = 11.84RR102 pKa = 11.84RR103 pKa = 11.84ATGKK107 pKa = 9.95LRR109 pKa = 11.84ARR111 pKa = 11.84RR112 pKa = 3.66
MM1 pKa = 6.98TNHH4 pKa = 5.74YY5 pKa = 7.38RR6 pKa = 11.84TCSRR10 pKa = 11.84LASGGQRR17 pKa = 11.84TASFPYY23 pKa = 9.84RR24 pKa = 11.84LAAVFFDD31 pKa = 3.5VSRR34 pKa = 11.84VSGVGLPTLLGICLLPLLLLCSGCGEE60 pKa = 4.5KK61 pKa = 10.79DD62 pKa = 3.06PAADD66 pKa = 4.52AEE68 pKa = 4.6VTTMKK73 pKa = 10.72ASAKK77 pKa = 9.08PEE79 pKa = 3.85PSAAEE84 pKa = 3.82IPLVTLHH91 pKa = 5.99TATRR95 pKa = 11.84GRR97 pKa = 11.84LPLRR101 pKa = 11.84RR102 pKa = 11.84RR103 pKa = 11.84ATGKK107 pKa = 9.95LRR109 pKa = 11.84ARR111 pKa = 11.84RR112 pKa = 3.66
Molecular weight: 12.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1692660 |
16 |
7041 |
389.9 |
43.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.458 ± 0.043 | 0.933 ± 0.023 |
6.226 ± 0.047 | 6.424 ± 0.034 |
4.607 ± 0.024 | 7.908 ± 0.039 |
1.786 ± 0.022 | 5.652 ± 0.029 |
4.411 ± 0.046 | 9.748 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.138 ± 0.017 | 4.49 ± 0.03 |
4.578 ± 0.03 | 3.367 ± 0.024 |
5.35 ± 0.048 | 6.109 ± 0.03 |
6.17 ± 0.058 | 6.785 ± 0.033 |
1.247 ± 0.015 | 3.615 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
