
Pleurotus ostreatus virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Betapartitivirus
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
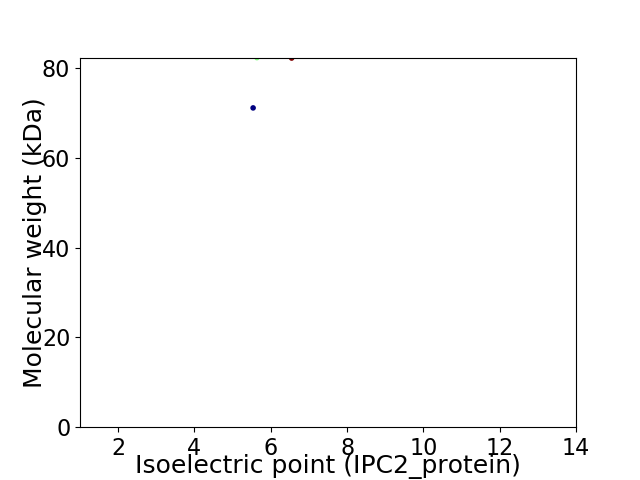
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q5GHF1|Q5GHF1_9VIRU RNA-dependent RNA polymerase OS=Pleurotus ostreatus virus 1 OX=674983 PE=4 SV=1
MM1 pKa = 7.41SSSAARR7 pKa = 11.84IQTASRR13 pKa = 11.84VAPAEE18 pKa = 3.71IRR20 pKa = 11.84TPPALAAIAKK30 pKa = 9.48KK31 pKa = 9.42FAPVILQDD39 pKa = 3.32NDD41 pKa = 3.63VPVPPSNADD50 pKa = 2.98YY51 pKa = 10.32FGRR54 pKa = 11.84MNLPTNIEE62 pKa = 4.12LDD64 pKa = 3.19SGNYY68 pKa = 9.79HH69 pKa = 6.72NIDD72 pKa = 3.33VQFDD76 pKa = 3.36TRR78 pKa = 11.84FILNYY83 pKa = 10.1IMYY86 pKa = 9.61HH87 pKa = 5.24VMNLAPEE94 pKa = 4.61LNFKK98 pKa = 9.16GHH100 pKa = 7.38PYY102 pKa = 10.3FSPLSYY108 pKa = 9.75IGYY111 pKa = 9.84CMHH114 pKa = 7.32LFYY117 pKa = 11.22ALLLACDD124 pKa = 3.74CTFRR128 pKa = 11.84SDD130 pKa = 3.43KK131 pKa = 10.66SYY133 pKa = 10.31HH134 pKa = 6.47ASRR137 pKa = 11.84FMTDD141 pKa = 2.99QEE143 pKa = 4.41RR144 pKa = 11.84KK145 pKa = 9.99DD146 pKa = 3.61LYY148 pKa = 10.86EE149 pKa = 4.05VLLNCQMPTFLSDD162 pKa = 4.55LFLEE166 pKa = 4.8LAPVYY171 pKa = 10.36DD172 pKa = 3.83PRR174 pKa = 11.84RR175 pKa = 11.84NNLLFVPSLAGHH187 pKa = 5.71SFEE190 pKa = 5.06HH191 pKa = 7.56DD192 pKa = 3.36YY193 pKa = 11.63GRR195 pKa = 11.84TLLPSMFYY203 pKa = 10.33AAHH206 pKa = 6.6HH207 pKa = 5.98MLASTRR213 pKa = 11.84TNKK216 pKa = 10.37DD217 pKa = 3.26PNDD220 pKa = 3.97VIDD223 pKa = 4.66DD224 pKa = 3.93CMALHH229 pKa = 7.04VITAGTTNFTISNYY243 pKa = 10.28LGTWYY248 pKa = 8.76ATGHH252 pKa = 6.29HH253 pKa = 7.03DD254 pKa = 3.12NWLNRR259 pKa = 11.84DD260 pKa = 3.55FLAFFNPLVGRR271 pKa = 11.84FLTQRR276 pKa = 11.84PTFARR281 pKa = 11.84MHH283 pKa = 6.69FEE285 pKa = 4.28TEE287 pKa = 3.96VLPIDD292 pKa = 4.02GTGNPYY298 pKa = 8.94TAFLLASDD306 pKa = 4.81EE307 pKa = 4.3NTSLATTVLTAPSTFVNANDD327 pKa = 3.91PKK329 pKa = 10.25SPKK332 pKa = 9.81LGSVLASLSGTLLLNYY348 pKa = 10.03AIEE351 pKa = 4.76PPTLPTWTAATYY363 pKa = 10.32NQDD366 pKa = 4.57DD367 pKa = 4.37NPSDD371 pKa = 3.69ISDD374 pKa = 3.3KK375 pKa = 11.1TFATEE380 pKa = 3.95HH381 pKa = 6.18NFLVDD386 pKa = 3.97EE387 pKa = 4.41STHH390 pKa = 7.07DD391 pKa = 3.45KK392 pKa = 9.91TLSYY396 pKa = 10.79PDD398 pKa = 5.49DD399 pKa = 4.01DD400 pKa = 4.44TGLNAAWYY408 pKa = 9.44AIAKK412 pKa = 8.65VKK414 pKa = 8.41HH415 pKa = 5.33TKK417 pKa = 10.21ARR419 pKa = 11.84TPFKK423 pKa = 10.73HH424 pKa = 6.87VIFNVKK430 pKa = 10.12DD431 pKa = 3.32HH432 pKa = 6.32LTPYY436 pKa = 10.45VLYY439 pKa = 8.92FQPYY443 pKa = 8.43DD444 pKa = 3.69VSPSSLDD451 pKa = 3.26LTIAAGIKK459 pKa = 9.41IEE461 pKa = 4.69HH462 pKa = 6.81GDD464 pKa = 3.17ISGFAINIEE473 pKa = 4.22QPDD476 pKa = 3.78SSLDD480 pKa = 3.69DD481 pKa = 3.73NNAQILQAAIRR492 pKa = 11.84LSKK495 pKa = 10.3VIRR498 pKa = 11.84VNAQTSAGDD507 pKa = 3.56NRR509 pKa = 11.84VSIAVRR515 pKa = 11.84EE516 pKa = 4.04PLDD519 pKa = 3.48RR520 pKa = 11.84TKK522 pKa = 10.89QGVMYY527 pKa = 10.44IFRR530 pKa = 11.84SLLKK534 pKa = 10.61SVFPVLDD541 pKa = 3.8NEE543 pKa = 4.69DD544 pKa = 3.59VKK546 pKa = 11.19STDD549 pKa = 2.93TVLPTNVGLTIEE561 pKa = 4.3RR562 pKa = 11.84GHH564 pKa = 6.48YY565 pKa = 10.86SFDD568 pKa = 2.97QGFNVKK574 pKa = 10.15AGSNGDD580 pKa = 3.34IKK582 pKa = 10.32TSDD585 pKa = 3.34DD586 pKa = 5.2SIYY589 pKa = 10.33LWSSYY594 pKa = 10.72RR595 pKa = 11.84YY596 pKa = 8.02VHH598 pKa = 6.49KK599 pKa = 10.6KK600 pKa = 10.09KK601 pKa = 10.65NPAPADD607 pKa = 3.24ISMLASLRR615 pKa = 11.84PFYY618 pKa = 11.07GNNVTLSRR626 pKa = 11.84SKK628 pKa = 11.12NPTLLIPHH636 pKa = 7.39
MM1 pKa = 7.41SSSAARR7 pKa = 11.84IQTASRR13 pKa = 11.84VAPAEE18 pKa = 3.71IRR20 pKa = 11.84TPPALAAIAKK30 pKa = 9.48KK31 pKa = 9.42FAPVILQDD39 pKa = 3.32NDD41 pKa = 3.63VPVPPSNADD50 pKa = 2.98YY51 pKa = 10.32FGRR54 pKa = 11.84MNLPTNIEE62 pKa = 4.12LDD64 pKa = 3.19SGNYY68 pKa = 9.79HH69 pKa = 6.72NIDD72 pKa = 3.33VQFDD76 pKa = 3.36TRR78 pKa = 11.84FILNYY83 pKa = 10.1IMYY86 pKa = 9.61HH87 pKa = 5.24VMNLAPEE94 pKa = 4.61LNFKK98 pKa = 9.16GHH100 pKa = 7.38PYY102 pKa = 10.3FSPLSYY108 pKa = 9.75IGYY111 pKa = 9.84CMHH114 pKa = 7.32LFYY117 pKa = 11.22ALLLACDD124 pKa = 3.74CTFRR128 pKa = 11.84SDD130 pKa = 3.43KK131 pKa = 10.66SYY133 pKa = 10.31HH134 pKa = 6.47ASRR137 pKa = 11.84FMTDD141 pKa = 2.99QEE143 pKa = 4.41RR144 pKa = 11.84KK145 pKa = 9.99DD146 pKa = 3.61LYY148 pKa = 10.86EE149 pKa = 4.05VLLNCQMPTFLSDD162 pKa = 4.55LFLEE166 pKa = 4.8LAPVYY171 pKa = 10.36DD172 pKa = 3.83PRR174 pKa = 11.84RR175 pKa = 11.84NNLLFVPSLAGHH187 pKa = 5.71SFEE190 pKa = 5.06HH191 pKa = 7.56DD192 pKa = 3.36YY193 pKa = 11.63GRR195 pKa = 11.84TLLPSMFYY203 pKa = 10.33AAHH206 pKa = 6.6HH207 pKa = 5.98MLASTRR213 pKa = 11.84TNKK216 pKa = 10.37DD217 pKa = 3.26PNDD220 pKa = 3.97VIDD223 pKa = 4.66DD224 pKa = 3.93CMALHH229 pKa = 7.04VITAGTTNFTISNYY243 pKa = 10.28LGTWYY248 pKa = 8.76ATGHH252 pKa = 6.29HH253 pKa = 7.03DD254 pKa = 3.12NWLNRR259 pKa = 11.84DD260 pKa = 3.55FLAFFNPLVGRR271 pKa = 11.84FLTQRR276 pKa = 11.84PTFARR281 pKa = 11.84MHH283 pKa = 6.69FEE285 pKa = 4.28TEE287 pKa = 3.96VLPIDD292 pKa = 4.02GTGNPYY298 pKa = 8.94TAFLLASDD306 pKa = 4.81EE307 pKa = 4.3NTSLATTVLTAPSTFVNANDD327 pKa = 3.91PKK329 pKa = 10.25SPKK332 pKa = 9.81LGSVLASLSGTLLLNYY348 pKa = 10.03AIEE351 pKa = 4.76PPTLPTWTAATYY363 pKa = 10.32NQDD366 pKa = 4.57DD367 pKa = 4.37NPSDD371 pKa = 3.69ISDD374 pKa = 3.3KK375 pKa = 11.1TFATEE380 pKa = 3.95HH381 pKa = 6.18NFLVDD386 pKa = 3.97EE387 pKa = 4.41STHH390 pKa = 7.07DD391 pKa = 3.45KK392 pKa = 9.91TLSYY396 pKa = 10.79PDD398 pKa = 5.49DD399 pKa = 4.01DD400 pKa = 4.44TGLNAAWYY408 pKa = 9.44AIAKK412 pKa = 8.65VKK414 pKa = 8.41HH415 pKa = 5.33TKK417 pKa = 10.21ARR419 pKa = 11.84TPFKK423 pKa = 10.73HH424 pKa = 6.87VIFNVKK430 pKa = 10.12DD431 pKa = 3.32HH432 pKa = 6.32LTPYY436 pKa = 10.45VLYY439 pKa = 8.92FQPYY443 pKa = 8.43DD444 pKa = 3.69VSPSSLDD451 pKa = 3.26LTIAAGIKK459 pKa = 9.41IEE461 pKa = 4.69HH462 pKa = 6.81GDD464 pKa = 3.17ISGFAINIEE473 pKa = 4.22QPDD476 pKa = 3.78SSLDD480 pKa = 3.69DD481 pKa = 3.73NNAQILQAAIRR492 pKa = 11.84LSKK495 pKa = 10.3VIRR498 pKa = 11.84VNAQTSAGDD507 pKa = 3.56NRR509 pKa = 11.84VSIAVRR515 pKa = 11.84EE516 pKa = 4.04PLDD519 pKa = 3.48RR520 pKa = 11.84TKK522 pKa = 10.89QGVMYY527 pKa = 10.44IFRR530 pKa = 11.84SLLKK534 pKa = 10.61SVFPVLDD541 pKa = 3.8NEE543 pKa = 4.69DD544 pKa = 3.59VKK546 pKa = 11.19STDD549 pKa = 2.93TVLPTNVGLTIEE561 pKa = 4.3RR562 pKa = 11.84GHH564 pKa = 6.48YY565 pKa = 10.86SFDD568 pKa = 2.97QGFNVKK574 pKa = 10.15AGSNGDD580 pKa = 3.34IKK582 pKa = 10.32TSDD585 pKa = 3.34DD586 pKa = 5.2SIYY589 pKa = 10.33LWSSYY594 pKa = 10.72RR595 pKa = 11.84YY596 pKa = 8.02VHH598 pKa = 6.49KK599 pKa = 10.6KK600 pKa = 10.09KK601 pKa = 10.65NPAPADD607 pKa = 3.24ISMLASLRR615 pKa = 11.84PFYY618 pKa = 11.07GNNVTLSRR626 pKa = 11.84SKK628 pKa = 11.12NPTLLIPHH636 pKa = 7.39
Molecular weight: 71.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5GHF1|Q5GHF1_9VIRU RNA-dependent RNA polymerase OS=Pleurotus ostreatus virus 1 OX=674983 PE=4 SV=1
MM1 pKa = 7.36SFLRR5 pKa = 11.84IRR7 pKa = 11.84DD8 pKa = 3.73YY9 pKa = 9.7FTEE12 pKa = 4.11RR13 pKa = 11.84LKK15 pKa = 11.09HH16 pKa = 6.71LSRR19 pKa = 11.84DD20 pKa = 3.03WKK22 pKa = 10.52IFQQSDD28 pKa = 3.47SDD30 pKa = 4.31PEE32 pKa = 4.34STLATHH38 pKa = 7.31LDD40 pKa = 3.3SDD42 pKa = 4.13IARR45 pKa = 11.84LEE47 pKa = 4.12HH48 pKa = 6.66GIKK51 pKa = 10.38SSLSDD56 pKa = 3.56DD57 pKa = 3.84QRR59 pKa = 11.84QQAYY63 pKa = 7.92EE64 pKa = 3.77RR65 pKa = 11.84EE66 pKa = 4.34YY67 pKa = 11.57NRR69 pKa = 11.84IHH71 pKa = 7.09SALHH75 pKa = 6.13DD76 pKa = 3.88KK77 pKa = 10.68ARR79 pKa = 11.84QDD81 pKa = 3.44GFPDD85 pKa = 2.95EE86 pKa = 5.17FYY88 pKa = 10.85RR89 pKa = 11.84SRR91 pKa = 11.84SIDD94 pKa = 3.76DD95 pKa = 4.56LPDD98 pKa = 3.2NRR100 pKa = 11.84IPPSGIIPLPYY111 pKa = 9.42EE112 pKa = 3.75YY113 pKa = 10.28HH114 pKa = 6.85RR115 pKa = 11.84SQVVISTEE123 pKa = 4.1EE124 pKa = 3.86VPEE127 pKa = 4.15TGFQIDD133 pKa = 3.67PRR135 pKa = 11.84IVRR138 pKa = 11.84ILRR141 pKa = 11.84NKK143 pKa = 8.39YY144 pKa = 8.56PQYY147 pKa = 10.38LPHH150 pKa = 5.07VTKK153 pKa = 10.7YY154 pKa = 10.38VRR156 pKa = 11.84PLGTTDD162 pKa = 2.98ATVKK166 pKa = 10.8DD167 pKa = 4.24FFKK170 pKa = 10.7PQIPSDD176 pKa = 4.99PIPEE180 pKa = 3.94ARR182 pKa = 11.84KK183 pKa = 9.59QRR185 pKa = 11.84ILDD188 pKa = 3.69LVISFLACTPFLPLHH203 pKa = 6.77FIDD206 pKa = 4.92SLWDD210 pKa = 3.28KK211 pKa = 10.8TPLHH215 pKa = 6.47TGTGYY220 pKa = 10.87FNRR223 pKa = 11.84HH224 pKa = 4.3SFAARR229 pKa = 11.84IHH231 pKa = 6.6AMFSAPRR238 pKa = 11.84LYY240 pKa = 10.58EE241 pKa = 3.7RR242 pKa = 11.84RR243 pKa = 11.84TTSKK247 pKa = 10.94GYY249 pKa = 9.87FINYY253 pKa = 8.87FLEE256 pKa = 4.42TARR259 pKa = 11.84STIHH263 pKa = 6.46NIKK266 pKa = 10.43LHH268 pKa = 6.1GFPFDD273 pKa = 3.74PSKK276 pKa = 11.58VPDD279 pKa = 4.11LGSALRR285 pKa = 11.84SFILKK290 pKa = 10.01RR291 pKa = 11.84PTMLFTRR298 pKa = 11.84NHH300 pKa = 5.58ISDD303 pKa = 3.44RR304 pKa = 11.84DD305 pKa = 3.78GNLKK309 pKa = 9.56QRR311 pKa = 11.84PVYY314 pKa = 11.14AMDD317 pKa = 4.44DD318 pKa = 3.43LFIRR322 pKa = 11.84LEE324 pKa = 4.1SMITFPLHH332 pKa = 5.13VMARR336 pKa = 11.84KK337 pKa = 9.6IEE339 pKa = 4.05CCIMYY344 pKa = 9.9GYY346 pKa = 8.32EE347 pKa = 4.21TIRR350 pKa = 11.84GSNRR354 pKa = 11.84QIDD357 pKa = 4.26KK358 pKa = 9.76IASSFRR364 pKa = 11.84SFFTIDD370 pKa = 2.61WSGFDD375 pKa = 3.15QRR377 pKa = 11.84LPRR380 pKa = 11.84VITDD384 pKa = 3.62IFWSDD389 pKa = 3.2FLEE392 pKa = 4.26RR393 pKa = 11.84MIVISHH399 pKa = 7.01GYY401 pKa = 8.47QPTYY405 pKa = 10.6DD406 pKa = 3.72YY407 pKa = 11.06PSYY410 pKa = 10.97PDD412 pKa = 3.52LTPDD416 pKa = 4.63KK417 pKa = 10.16MFQRR421 pKa = 11.84MDD423 pKa = 3.78NILFFLHH430 pKa = 5.31TWYY433 pKa = 11.09NNLVFVTADD442 pKa = 2.98GFAYY446 pKa = 9.65IRR448 pKa = 11.84TCAGVPSGLLNTQYY462 pKa = 11.45LDD464 pKa = 3.46SFSNLFLIFDD474 pKa = 3.96GLIEE478 pKa = 4.56FGCSDD483 pKa = 3.66AEE485 pKa = 4.06IYY487 pKa = 10.65QIFLLVMGDD496 pKa = 3.81DD497 pKa = 3.92NSAFTLWSIAKK508 pKa = 10.1LEE510 pKa = 4.09EE511 pKa = 3.95FLSFFEE517 pKa = 4.73SYY519 pKa = 10.89ALRR522 pKa = 11.84RR523 pKa = 11.84FGMVLSKK530 pKa = 9.28TKK532 pKa = 10.75SVITVIRR539 pKa = 11.84GKK541 pKa = 9.98IEE543 pKa = 3.88TLSYY547 pKa = 8.71QCNYY551 pKa = 9.43GAPKK555 pKa = 10.23RR556 pKa = 11.84PLAKK560 pKa = 10.21LVAQLCYY567 pKa = 10.0PEE569 pKa = 4.84RR570 pKa = 11.84GPRR573 pKa = 11.84AKK575 pKa = 9.4YY576 pKa = 8.69TSARR580 pKa = 11.84AIGMAYY586 pKa = 9.68AACAMDD592 pKa = 3.87RR593 pKa = 11.84TFHH596 pKa = 6.75DD597 pKa = 4.25LCRR600 pKa = 11.84DD601 pKa = 3.19IYY603 pKa = 11.45YY604 pKa = 10.74EE605 pKa = 4.17FLDD608 pKa = 5.01DD609 pKa = 4.66SASPDD614 pKa = 3.17EE615 pKa = 4.52PFFFEE620 pKa = 5.15HH621 pKa = 5.37VQAYY625 pKa = 10.17LPGILRR631 pKa = 11.84TDD633 pKa = 3.51EE634 pKa = 4.25SLSTQISLSSFPSFLTVQQHH654 pKa = 4.89ISRR657 pKa = 11.84WQGPLSYY664 pKa = 10.45YY665 pKa = 9.65PKK667 pKa = 9.13WDD669 pKa = 3.4RR670 pKa = 11.84AHH672 pKa = 7.35FINDD676 pKa = 3.65PDD678 pKa = 4.36VIPPSAEE685 pKa = 3.34TMAEE689 pKa = 3.88YY690 pKa = 10.32RR691 pKa = 11.84SRR693 pKa = 11.84NSIPRR698 pKa = 11.84RR699 pKa = 11.84DD700 pKa = 4.36IPSLWQQ706 pKa = 3.32
MM1 pKa = 7.36SFLRR5 pKa = 11.84IRR7 pKa = 11.84DD8 pKa = 3.73YY9 pKa = 9.7FTEE12 pKa = 4.11RR13 pKa = 11.84LKK15 pKa = 11.09HH16 pKa = 6.71LSRR19 pKa = 11.84DD20 pKa = 3.03WKK22 pKa = 10.52IFQQSDD28 pKa = 3.47SDD30 pKa = 4.31PEE32 pKa = 4.34STLATHH38 pKa = 7.31LDD40 pKa = 3.3SDD42 pKa = 4.13IARR45 pKa = 11.84LEE47 pKa = 4.12HH48 pKa = 6.66GIKK51 pKa = 10.38SSLSDD56 pKa = 3.56DD57 pKa = 3.84QRR59 pKa = 11.84QQAYY63 pKa = 7.92EE64 pKa = 3.77RR65 pKa = 11.84EE66 pKa = 4.34YY67 pKa = 11.57NRR69 pKa = 11.84IHH71 pKa = 7.09SALHH75 pKa = 6.13DD76 pKa = 3.88KK77 pKa = 10.68ARR79 pKa = 11.84QDD81 pKa = 3.44GFPDD85 pKa = 2.95EE86 pKa = 5.17FYY88 pKa = 10.85RR89 pKa = 11.84SRR91 pKa = 11.84SIDD94 pKa = 3.76DD95 pKa = 4.56LPDD98 pKa = 3.2NRR100 pKa = 11.84IPPSGIIPLPYY111 pKa = 9.42EE112 pKa = 3.75YY113 pKa = 10.28HH114 pKa = 6.85RR115 pKa = 11.84SQVVISTEE123 pKa = 4.1EE124 pKa = 3.86VPEE127 pKa = 4.15TGFQIDD133 pKa = 3.67PRR135 pKa = 11.84IVRR138 pKa = 11.84ILRR141 pKa = 11.84NKK143 pKa = 8.39YY144 pKa = 8.56PQYY147 pKa = 10.38LPHH150 pKa = 5.07VTKK153 pKa = 10.7YY154 pKa = 10.38VRR156 pKa = 11.84PLGTTDD162 pKa = 2.98ATVKK166 pKa = 10.8DD167 pKa = 4.24FFKK170 pKa = 10.7PQIPSDD176 pKa = 4.99PIPEE180 pKa = 3.94ARR182 pKa = 11.84KK183 pKa = 9.59QRR185 pKa = 11.84ILDD188 pKa = 3.69LVISFLACTPFLPLHH203 pKa = 6.77FIDD206 pKa = 4.92SLWDD210 pKa = 3.28KK211 pKa = 10.8TPLHH215 pKa = 6.47TGTGYY220 pKa = 10.87FNRR223 pKa = 11.84HH224 pKa = 4.3SFAARR229 pKa = 11.84IHH231 pKa = 6.6AMFSAPRR238 pKa = 11.84LYY240 pKa = 10.58EE241 pKa = 3.7RR242 pKa = 11.84RR243 pKa = 11.84TTSKK247 pKa = 10.94GYY249 pKa = 9.87FINYY253 pKa = 8.87FLEE256 pKa = 4.42TARR259 pKa = 11.84STIHH263 pKa = 6.46NIKK266 pKa = 10.43LHH268 pKa = 6.1GFPFDD273 pKa = 3.74PSKK276 pKa = 11.58VPDD279 pKa = 4.11LGSALRR285 pKa = 11.84SFILKK290 pKa = 10.01RR291 pKa = 11.84PTMLFTRR298 pKa = 11.84NHH300 pKa = 5.58ISDD303 pKa = 3.44RR304 pKa = 11.84DD305 pKa = 3.78GNLKK309 pKa = 9.56QRR311 pKa = 11.84PVYY314 pKa = 11.14AMDD317 pKa = 4.44DD318 pKa = 3.43LFIRR322 pKa = 11.84LEE324 pKa = 4.1SMITFPLHH332 pKa = 5.13VMARR336 pKa = 11.84KK337 pKa = 9.6IEE339 pKa = 4.05CCIMYY344 pKa = 9.9GYY346 pKa = 8.32EE347 pKa = 4.21TIRR350 pKa = 11.84GSNRR354 pKa = 11.84QIDD357 pKa = 4.26KK358 pKa = 9.76IASSFRR364 pKa = 11.84SFFTIDD370 pKa = 2.61WSGFDD375 pKa = 3.15QRR377 pKa = 11.84LPRR380 pKa = 11.84VITDD384 pKa = 3.62IFWSDD389 pKa = 3.2FLEE392 pKa = 4.26RR393 pKa = 11.84MIVISHH399 pKa = 7.01GYY401 pKa = 8.47QPTYY405 pKa = 10.6DD406 pKa = 3.72YY407 pKa = 11.06PSYY410 pKa = 10.97PDD412 pKa = 3.52LTPDD416 pKa = 4.63KK417 pKa = 10.16MFQRR421 pKa = 11.84MDD423 pKa = 3.78NILFFLHH430 pKa = 5.31TWYY433 pKa = 11.09NNLVFVTADD442 pKa = 2.98GFAYY446 pKa = 9.65IRR448 pKa = 11.84TCAGVPSGLLNTQYY462 pKa = 11.45LDD464 pKa = 3.46SFSNLFLIFDD474 pKa = 3.96GLIEE478 pKa = 4.56FGCSDD483 pKa = 3.66AEE485 pKa = 4.06IYY487 pKa = 10.65QIFLLVMGDD496 pKa = 3.81DD497 pKa = 3.92NSAFTLWSIAKK508 pKa = 10.1LEE510 pKa = 4.09EE511 pKa = 3.95FLSFFEE517 pKa = 4.73SYY519 pKa = 10.89ALRR522 pKa = 11.84RR523 pKa = 11.84FGMVLSKK530 pKa = 9.28TKK532 pKa = 10.75SVITVIRR539 pKa = 11.84GKK541 pKa = 9.98IEE543 pKa = 3.88TLSYY547 pKa = 8.71QCNYY551 pKa = 9.43GAPKK555 pKa = 10.23RR556 pKa = 11.84PLAKK560 pKa = 10.21LVAQLCYY567 pKa = 10.0PEE569 pKa = 4.84RR570 pKa = 11.84GPRR573 pKa = 11.84AKK575 pKa = 9.4YY576 pKa = 8.69TSARR580 pKa = 11.84AIGMAYY586 pKa = 9.68AACAMDD592 pKa = 3.87RR593 pKa = 11.84TFHH596 pKa = 6.75DD597 pKa = 4.25LCRR600 pKa = 11.84DD601 pKa = 3.19IYY603 pKa = 11.45YY604 pKa = 10.74EE605 pKa = 4.17FLDD608 pKa = 5.01DD609 pKa = 4.66SASPDD614 pKa = 3.17EE615 pKa = 4.52PFFFEE620 pKa = 5.15HH621 pKa = 5.37VQAYY625 pKa = 10.17LPGILRR631 pKa = 11.84TDD633 pKa = 3.51EE634 pKa = 4.25SLSTQISLSSFPSFLTVQQHH654 pKa = 4.89ISRR657 pKa = 11.84WQGPLSYY664 pKa = 10.45YY665 pKa = 9.65PKK667 pKa = 9.13WDD669 pKa = 3.4RR670 pKa = 11.84AHH672 pKa = 7.35FINDD676 pKa = 3.65PDD678 pKa = 4.36VIPPSAEE685 pKa = 3.34TMAEE689 pKa = 3.88YY690 pKa = 10.32RR691 pKa = 11.84SRR693 pKa = 11.84NSIPRR698 pKa = 11.84RR699 pKa = 11.84DD700 pKa = 4.36IPSLWQQ706 pKa = 3.32
Molecular weight: 82.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1342 |
636 |
706 |
671.0 |
76.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.855 ± 0.975 | 1.043 ± 0.17 |
7.526 ± 0.014 | 3.577 ± 0.493 |
6.334 ± 0.652 | 4.024 ± 0.042 |
3.204 ± 0.168 | 6.557 ± 0.903 |
3.875 ± 0.141 | 9.538 ± 0.45 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.086 ± 0.028 | 4.471 ± 1.407 |
6.632 ± 0.019 | 2.981 ± 0.514 |
6.185 ± 1.28 | 8.271 ± 0.27 |
6.557 ± 0.757 | 4.396 ± 0.73 |
1.043 ± 0.17 | 4.844 ± 0.187 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
