
Lunatimonas lonarensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cyclobacteriaceae; Lunatimonas
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
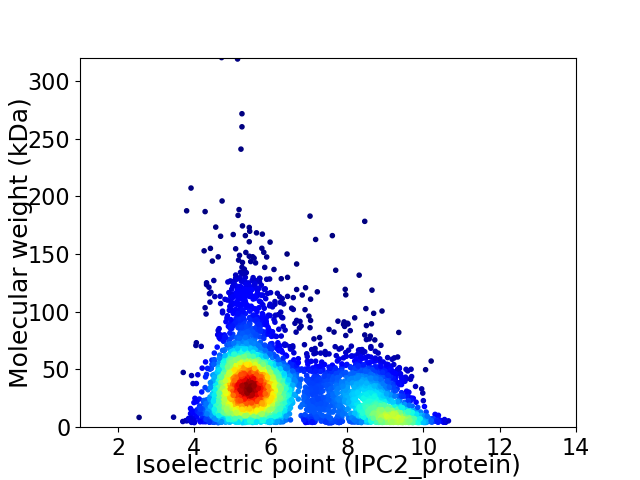
Virtual 2D-PAGE plot for 4825 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7ZS84|R7ZS84_9BACT Alpha-L-rhamnosidase OS=Lunatimonas lonarensis OX=1232681 GN=ADIS_2735 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 10.39SLSIYY7 pKa = 9.14INHH10 pKa = 7.52GIFVLIAMVLLFFDD24 pKa = 4.42FQAAKK29 pKa = 10.62AQGFNNNEE37 pKa = 4.08WIFGYY42 pKa = 10.69CEE44 pKa = 4.08GGEE47 pKa = 4.01NNYY50 pKa = 10.64LSFGKK55 pKa = 10.28DD56 pKa = 2.56GTARR60 pKa = 11.84VRR62 pKa = 11.84TLSPSVVLGKK72 pKa = 11.01GNTAMAIDD80 pKa = 5.62PISGDD85 pKa = 3.01ILFYY89 pKa = 10.83TDD91 pKa = 3.45GALVYY96 pKa = 10.49NYY98 pKa = 10.02LNQPMQGIIGEE109 pKa = 4.31LGGDD113 pKa = 3.22EE114 pKa = 4.42TGRR117 pKa = 11.84QTVAIAVQEE126 pKa = 3.95FDD128 pKa = 5.22PIPGGSRR135 pKa = 11.84SFYY138 pKa = 10.62LFFINRR144 pKa = 11.84RR145 pKa = 11.84GEE147 pKa = 3.96LEE149 pKa = 3.96YY150 pKa = 11.19DD151 pKa = 3.62VVDD154 pKa = 3.99MNNQGGAPASQPPAGAVSGGGILGTAQGGVAVIKK188 pKa = 9.71TPASEE193 pKa = 4.0SHH195 pKa = 6.91LVFFDD200 pKa = 3.32NGNLQVYY207 pKa = 10.03RR208 pKa = 11.84IEE210 pKa = 4.51DD211 pKa = 3.27QLGQFTLLQSEE222 pKa = 4.6ALDD225 pKa = 3.7FVPEE229 pKa = 3.99AFVFDD234 pKa = 3.95EE235 pKa = 4.8GSEE238 pKa = 4.1KK239 pKa = 10.55FVVIPQGTGADD250 pKa = 3.93LIVFDD255 pKa = 5.59FDD257 pKa = 4.27ASSGTLGNAQTLEE270 pKa = 4.21NTGGDD275 pKa = 3.6DD276 pKa = 4.58PIFGVAFSPDD286 pKa = 2.65GSMLYY291 pKa = 10.4FSRR294 pKa = 11.84GNQLFRR300 pKa = 11.84IGADD304 pKa = 3.09GSLADD309 pKa = 4.34EE310 pKa = 4.65LPEE313 pKa = 4.12EE314 pKa = 4.46EE315 pKa = 4.75EE316 pKa = 4.98EE317 pKa = 4.27EE318 pKa = 4.21DD319 pKa = 5.6QEE321 pKa = 5.96DD322 pKa = 4.28EE323 pKa = 4.72DD324 pKa = 5.09PEE326 pKa = 4.43ILTGPVEE333 pKa = 4.2VALGHH338 pKa = 6.43DD339 pKa = 4.3LFRR342 pKa = 11.84IYY344 pKa = 10.8DD345 pKa = 3.77LRR347 pKa = 11.84VGPDD351 pKa = 3.03GQLYY355 pKa = 9.76YY356 pKa = 9.97IYY358 pKa = 10.04EE359 pKa = 4.14EE360 pKa = 4.71TEE362 pKa = 3.92GGPQFLGRR370 pKa = 11.84IVNPDD375 pKa = 3.15EE376 pKa = 4.21EE377 pKa = 4.89ALGEE381 pKa = 4.22LTVEE385 pKa = 4.48ADD387 pKa = 3.52PFEE390 pKa = 4.49GTDD393 pKa = 3.44FCGRR397 pKa = 11.84VFTVFTPTIDD407 pKa = 3.22VGPEE411 pKa = 3.35VDD413 pKa = 4.75FTWEE417 pKa = 3.92PFDD420 pKa = 3.8PCMNNPLLLTSQISPPNYY438 pKa = 9.54RR439 pKa = 11.84PVSFNWEE446 pKa = 3.91ILPPLVDD453 pKa = 3.67EE454 pKa = 5.24DD455 pKa = 4.42GEE457 pKa = 4.37EE458 pKa = 4.42IEE460 pKa = 4.39MDD462 pKa = 3.64LSKK465 pKa = 11.13EE466 pKa = 3.9HH467 pKa = 6.81LLLPRR472 pKa = 11.84EE473 pKa = 4.48ATSEE477 pKa = 4.04QQVTVVLTVTFANGEE492 pKa = 4.28TRR494 pKa = 11.84EE495 pKa = 4.12VTQTINFQEE504 pKa = 4.43NDD506 pKa = 3.52LQVQFNPADD515 pKa = 3.66TTLCDD520 pKa = 3.71PACIDD525 pKa = 4.78LMPLVDD531 pKa = 3.86VQSGQDD537 pKa = 3.07QGGQPGQPGVGQPGFGQPGFGQPGFGQPGFGQPGVGQPGFGQQPGQGGNYY587 pKa = 9.81EE588 pKa = 4.32YY589 pKa = 10.67FWSNKK594 pKa = 9.62RR595 pKa = 11.84EE596 pKa = 4.29DD597 pKa = 3.47GWGPEE602 pKa = 4.27APNEE606 pKa = 4.19VCRR609 pKa = 11.84PGYY612 pKa = 8.9YY613 pKa = 8.62WVLVRR618 pKa = 11.84EE619 pKa = 4.47VGSTCYY625 pKa = 10.18SYY627 pKa = 11.6AGIRR631 pKa = 11.84IQMWDD636 pKa = 3.53VQDD639 pKa = 3.35QTNNIWYY646 pKa = 9.24FGDD649 pKa = 3.94GAGLDD654 pKa = 4.29FNPDD658 pKa = 3.48PDD660 pKa = 5.66DD661 pKa = 4.66PDD663 pKa = 5.29APVPRR668 pKa = 11.84PIPSRR673 pKa = 11.84HH674 pKa = 6.05PRR676 pKa = 11.84NIPAGVTTVSDD687 pKa = 3.22QAGQVLFFTDD697 pKa = 5.3GEE699 pKa = 4.97TVWDD703 pKa = 4.1LNGNPMQNGLDD714 pKa = 3.39IGGDD718 pKa = 3.68PQSAGSVIAVPVASDD733 pKa = 3.13EE734 pKa = 4.34TLFYY738 pKa = 11.15LFTTRR743 pKa = 11.84RR744 pKa = 11.84GADD747 pKa = 3.36GNSVVSFSMVDD758 pKa = 2.94IKK760 pKa = 11.46GNNDD764 pKa = 3.45DD765 pKa = 3.93GVGNVVTKK773 pKa = 11.23NNFLFSSSTEE783 pKa = 3.53HH784 pKa = 6.88SAALNAGDD792 pKa = 3.95TTWVAFHH799 pKa = 6.73EE800 pKa = 4.75AGNNTFRR807 pKa = 11.84MYY809 pKa = 10.17PVSGQGIGQPVFSSVGGVHH828 pKa = 6.58SFGSSSGVMKK838 pKa = 10.48FSGDD842 pKa = 3.42GSRR845 pKa = 11.84LAITFQEE852 pKa = 4.07GGMNKK857 pKa = 9.81LQIFDD862 pKa = 4.25FDD864 pKa = 4.11QNTGEE869 pKa = 4.11LTEE872 pKa = 4.6FALLDD877 pKa = 4.31LGTDD881 pKa = 3.3GDD883 pKa = 4.41VYY885 pKa = 11.32GLEE888 pKa = 4.12FSTDD892 pKa = 2.93NTRR895 pKa = 11.84VYY897 pKa = 10.65VSYY900 pKa = 11.04RR901 pKa = 11.84NGGPGVEE908 pKa = 4.04EE909 pKa = 5.07FYY911 pKa = 10.99LSSVEE916 pKa = 4.15EE917 pKa = 4.41TDD919 pKa = 6.09DD920 pKa = 4.26SDD922 pKa = 4.03PDD924 pKa = 3.66NPVTTRR930 pKa = 11.84CQEE933 pKa = 4.14CFDD936 pKa = 4.03SAVEE940 pKa = 3.82RR941 pKa = 11.84RR942 pKa = 11.84EE943 pKa = 3.74IEE945 pKa = 3.96QCILASRR952 pKa = 11.84AAISQTGGLPLGALQVGPDD971 pKa = 3.56GQIYY975 pKa = 9.1VAVVGSNQIGQILPGRR991 pKa = 11.84GCNPSTFNQDD1001 pKa = 2.41AVEE1004 pKa = 4.11PMPGTTNLGLPSFVQQSGSNIPEE1027 pKa = 4.2PSLAGPNRR1035 pKa = 11.84LCVGADD1041 pKa = 3.34EE1042 pKa = 4.79GAVGLFEE1049 pKa = 5.87GGGEE1053 pKa = 4.09PDD1055 pKa = 2.89IDD1057 pKa = 4.14LYY1059 pKa = 11.62NWTIFNEE1066 pKa = 3.94QGEE1069 pKa = 4.55IVDD1072 pKa = 3.61QFLNGAEE1079 pKa = 3.9EE1080 pKa = 4.18LQNLEE1085 pKa = 4.16YY1086 pKa = 10.91QFATAGLFTVQLEE1099 pKa = 4.4VNRR1102 pKa = 11.84CGEE1105 pKa = 3.81PWDD1108 pKa = 3.98EE1109 pKa = 4.08IFRR1112 pKa = 11.84LEE1114 pKa = 4.2VEE1116 pKa = 4.71VVDD1119 pKa = 4.16SPEE1122 pKa = 5.24IILPTDD1128 pKa = 2.99IALCADD1134 pKa = 3.58AVLEE1138 pKa = 4.26LVAVDD1143 pKa = 4.25PDD1145 pKa = 3.84DD1146 pKa = 4.38PRR1148 pKa = 11.84LDD1150 pKa = 3.5EE1151 pKa = 5.85YY1152 pKa = 11.6VFEE1155 pKa = 4.46WVNAAGEE1162 pKa = 4.32IVGDD1166 pKa = 3.83QNVLEE1171 pKa = 4.27VTEE1174 pKa = 4.07EE1175 pKa = 3.96SIYY1178 pKa = 10.55TVTVAYY1184 pKa = 9.89RR1185 pKa = 11.84LPEE1188 pKa = 4.45DD1189 pKa = 4.62ADD1191 pKa = 3.42PEE1193 pKa = 4.3MFQTCPVSRR1202 pKa = 11.84SVFVGPPFDD1211 pKa = 4.63FDD1213 pKa = 3.43IDD1215 pKa = 3.62QSEE1218 pKa = 4.53EE1219 pKa = 4.2EE1220 pKa = 4.42VCLGEE1225 pKa = 4.41TVTFTPNSPVSGSWSIHH1242 pKa = 5.92RR1243 pKa = 11.84EE1244 pKa = 3.8GSQDD1248 pKa = 4.1RR1249 pKa = 11.84ISLPDD1254 pKa = 3.66TLQLTLNTAILGEE1267 pKa = 4.38AGDD1270 pKa = 4.02YY1271 pKa = 11.14EE1272 pKa = 5.6LFFLTSDD1279 pKa = 4.33PLDD1282 pKa = 3.64EE1283 pKa = 4.46TCLFEE1288 pKa = 5.45RR1289 pKa = 11.84SAFLRR1294 pKa = 11.84VNAGPQFEE1302 pKa = 5.06VIATSPAQSCEE1313 pKa = 3.65IGDD1316 pKa = 3.99GVLEE1320 pKa = 4.13IQAGVLLDD1328 pKa = 3.6SLVVQEE1334 pKa = 4.32TGQLFFDD1341 pKa = 4.36VLPNDD1346 pKa = 4.19IIVVDD1351 pKa = 4.08GLEE1354 pKa = 4.06PGIYY1358 pKa = 9.45TLLGFANGCSSSQAAVIEE1376 pKa = 4.3NANPPSDD1383 pKa = 3.51VLFEE1387 pKa = 4.42LEE1389 pKa = 4.75TRR1391 pKa = 11.84DD1392 pKa = 3.74EE1393 pKa = 4.14QCAPEE1398 pKa = 4.24GLVAGGISILFGNGPASGVYY1418 pKa = 9.31EE1419 pKa = 4.17VLGLDD1424 pKa = 3.24TGEE1427 pKa = 5.05LISEE1431 pKa = 4.36SFANQSVIALDD1442 pKa = 4.23LPPGRR1447 pKa = 11.84YY1448 pKa = 8.73LVSVTDD1454 pKa = 3.44VGGCEE1459 pKa = 3.85VPAPSEE1465 pKa = 4.18VIVNEE1470 pKa = 3.9FSEE1473 pKa = 4.59SVEE1476 pKa = 4.05VEE1478 pKa = 3.97LFSPNLCGPVVSTSITASGDD1498 pKa = 3.16FLAVDD1503 pKa = 3.88RR1504 pKa = 11.84FGWFRR1509 pKa = 11.84EE1510 pKa = 4.12VNGEE1514 pKa = 4.35FIPITGEE1521 pKa = 3.6SGMVISVSEE1530 pKa = 3.94PGRR1533 pKa = 11.84YY1534 pKa = 7.73EE1535 pKa = 3.54VRR1537 pKa = 11.84LYY1539 pKa = 11.47NEE1541 pKa = 4.49FDD1543 pKa = 4.31CIIGSGVIDD1552 pKa = 4.45VIQSATEE1559 pKa = 3.88PPVLSPRR1566 pKa = 11.84YY1567 pKa = 9.31VICTLDD1573 pKa = 3.45GNLAEE1578 pKa = 4.57LQAGAGFASYY1588 pKa = 10.1SWSLDD1593 pKa = 3.16GQEE1596 pKa = 4.77LGRR1599 pKa = 11.84EE1600 pKa = 4.14ARR1602 pKa = 11.84FVPTLPGRR1610 pKa = 11.84YY1611 pKa = 8.48EE1612 pKa = 4.04LLVTDD1617 pKa = 3.95EE1618 pKa = 4.94AGCEE1622 pKa = 3.8FLVDD1626 pKa = 4.36FEE1628 pKa = 4.76VEE1630 pKa = 4.0EE1631 pKa = 4.39DD1632 pKa = 3.73CEE1634 pKa = 4.63INVRR1638 pKa = 11.84FPNAIIPGSPTRR1650 pKa = 11.84NFLLYY1655 pKa = 10.24TRR1657 pKa = 11.84GPIDD1661 pKa = 3.23KK1662 pKa = 10.29LGVYY1666 pKa = 9.74IYY1668 pKa = 10.62NRR1670 pKa = 11.84WGEE1673 pKa = 4.05LIYY1676 pKa = 11.0YY1677 pKa = 10.01CEE1679 pKa = 4.01EE1680 pKa = 4.15TNPSEE1685 pKa = 4.55EE1686 pKa = 4.37ISICTWDD1693 pKa = 4.26GYY1695 pKa = 11.74VNGQKK1700 pKa = 10.13VPVGTYY1706 pKa = 8.52PVVVKK1711 pKa = 9.01FSNQQQQIEE1720 pKa = 4.32RR1721 pKa = 11.84TIRR1724 pKa = 11.84RR1725 pKa = 11.84AIVVIEE1731 pKa = 3.9
MM1 pKa = 7.67KK2 pKa = 10.39SLSIYY7 pKa = 9.14INHH10 pKa = 7.52GIFVLIAMVLLFFDD24 pKa = 4.42FQAAKK29 pKa = 10.62AQGFNNNEE37 pKa = 4.08WIFGYY42 pKa = 10.69CEE44 pKa = 4.08GGEE47 pKa = 4.01NNYY50 pKa = 10.64LSFGKK55 pKa = 10.28DD56 pKa = 2.56GTARR60 pKa = 11.84VRR62 pKa = 11.84TLSPSVVLGKK72 pKa = 11.01GNTAMAIDD80 pKa = 5.62PISGDD85 pKa = 3.01ILFYY89 pKa = 10.83TDD91 pKa = 3.45GALVYY96 pKa = 10.49NYY98 pKa = 10.02LNQPMQGIIGEE109 pKa = 4.31LGGDD113 pKa = 3.22EE114 pKa = 4.42TGRR117 pKa = 11.84QTVAIAVQEE126 pKa = 3.95FDD128 pKa = 5.22PIPGGSRR135 pKa = 11.84SFYY138 pKa = 10.62LFFINRR144 pKa = 11.84RR145 pKa = 11.84GEE147 pKa = 3.96LEE149 pKa = 3.96YY150 pKa = 11.19DD151 pKa = 3.62VVDD154 pKa = 3.99MNNQGGAPASQPPAGAVSGGGILGTAQGGVAVIKK188 pKa = 9.71TPASEE193 pKa = 4.0SHH195 pKa = 6.91LVFFDD200 pKa = 3.32NGNLQVYY207 pKa = 10.03RR208 pKa = 11.84IEE210 pKa = 4.51DD211 pKa = 3.27QLGQFTLLQSEE222 pKa = 4.6ALDD225 pKa = 3.7FVPEE229 pKa = 3.99AFVFDD234 pKa = 3.95EE235 pKa = 4.8GSEE238 pKa = 4.1KK239 pKa = 10.55FVVIPQGTGADD250 pKa = 3.93LIVFDD255 pKa = 5.59FDD257 pKa = 4.27ASSGTLGNAQTLEE270 pKa = 4.21NTGGDD275 pKa = 3.6DD276 pKa = 4.58PIFGVAFSPDD286 pKa = 2.65GSMLYY291 pKa = 10.4FSRR294 pKa = 11.84GNQLFRR300 pKa = 11.84IGADD304 pKa = 3.09GSLADD309 pKa = 4.34EE310 pKa = 4.65LPEE313 pKa = 4.12EE314 pKa = 4.46EE315 pKa = 4.75EE316 pKa = 4.98EE317 pKa = 4.27EE318 pKa = 4.21DD319 pKa = 5.6QEE321 pKa = 5.96DD322 pKa = 4.28EE323 pKa = 4.72DD324 pKa = 5.09PEE326 pKa = 4.43ILTGPVEE333 pKa = 4.2VALGHH338 pKa = 6.43DD339 pKa = 4.3LFRR342 pKa = 11.84IYY344 pKa = 10.8DD345 pKa = 3.77LRR347 pKa = 11.84VGPDD351 pKa = 3.03GQLYY355 pKa = 9.76YY356 pKa = 9.97IYY358 pKa = 10.04EE359 pKa = 4.14EE360 pKa = 4.71TEE362 pKa = 3.92GGPQFLGRR370 pKa = 11.84IVNPDD375 pKa = 3.15EE376 pKa = 4.21EE377 pKa = 4.89ALGEE381 pKa = 4.22LTVEE385 pKa = 4.48ADD387 pKa = 3.52PFEE390 pKa = 4.49GTDD393 pKa = 3.44FCGRR397 pKa = 11.84VFTVFTPTIDD407 pKa = 3.22VGPEE411 pKa = 3.35VDD413 pKa = 4.75FTWEE417 pKa = 3.92PFDD420 pKa = 3.8PCMNNPLLLTSQISPPNYY438 pKa = 9.54RR439 pKa = 11.84PVSFNWEE446 pKa = 3.91ILPPLVDD453 pKa = 3.67EE454 pKa = 5.24DD455 pKa = 4.42GEE457 pKa = 4.37EE458 pKa = 4.42IEE460 pKa = 4.39MDD462 pKa = 3.64LSKK465 pKa = 11.13EE466 pKa = 3.9HH467 pKa = 6.81LLLPRR472 pKa = 11.84EE473 pKa = 4.48ATSEE477 pKa = 4.04QQVTVVLTVTFANGEE492 pKa = 4.28TRR494 pKa = 11.84EE495 pKa = 4.12VTQTINFQEE504 pKa = 4.43NDD506 pKa = 3.52LQVQFNPADD515 pKa = 3.66TTLCDD520 pKa = 3.71PACIDD525 pKa = 4.78LMPLVDD531 pKa = 3.86VQSGQDD537 pKa = 3.07QGGQPGQPGVGQPGFGQPGFGQPGFGQPGFGQPGVGQPGFGQQPGQGGNYY587 pKa = 9.81EE588 pKa = 4.32YY589 pKa = 10.67FWSNKK594 pKa = 9.62RR595 pKa = 11.84EE596 pKa = 4.29DD597 pKa = 3.47GWGPEE602 pKa = 4.27APNEE606 pKa = 4.19VCRR609 pKa = 11.84PGYY612 pKa = 8.9YY613 pKa = 8.62WVLVRR618 pKa = 11.84EE619 pKa = 4.47VGSTCYY625 pKa = 10.18SYY627 pKa = 11.6AGIRR631 pKa = 11.84IQMWDD636 pKa = 3.53VQDD639 pKa = 3.35QTNNIWYY646 pKa = 9.24FGDD649 pKa = 3.94GAGLDD654 pKa = 4.29FNPDD658 pKa = 3.48PDD660 pKa = 5.66DD661 pKa = 4.66PDD663 pKa = 5.29APVPRR668 pKa = 11.84PIPSRR673 pKa = 11.84HH674 pKa = 6.05PRR676 pKa = 11.84NIPAGVTTVSDD687 pKa = 3.22QAGQVLFFTDD697 pKa = 5.3GEE699 pKa = 4.97TVWDD703 pKa = 4.1LNGNPMQNGLDD714 pKa = 3.39IGGDD718 pKa = 3.68PQSAGSVIAVPVASDD733 pKa = 3.13EE734 pKa = 4.34TLFYY738 pKa = 11.15LFTTRR743 pKa = 11.84RR744 pKa = 11.84GADD747 pKa = 3.36GNSVVSFSMVDD758 pKa = 2.94IKK760 pKa = 11.46GNNDD764 pKa = 3.45DD765 pKa = 3.93GVGNVVTKK773 pKa = 11.23NNFLFSSSTEE783 pKa = 3.53HH784 pKa = 6.88SAALNAGDD792 pKa = 3.95TTWVAFHH799 pKa = 6.73EE800 pKa = 4.75AGNNTFRR807 pKa = 11.84MYY809 pKa = 10.17PVSGQGIGQPVFSSVGGVHH828 pKa = 6.58SFGSSSGVMKK838 pKa = 10.48FSGDD842 pKa = 3.42GSRR845 pKa = 11.84LAITFQEE852 pKa = 4.07GGMNKK857 pKa = 9.81LQIFDD862 pKa = 4.25FDD864 pKa = 4.11QNTGEE869 pKa = 4.11LTEE872 pKa = 4.6FALLDD877 pKa = 4.31LGTDD881 pKa = 3.3GDD883 pKa = 4.41VYY885 pKa = 11.32GLEE888 pKa = 4.12FSTDD892 pKa = 2.93NTRR895 pKa = 11.84VYY897 pKa = 10.65VSYY900 pKa = 11.04RR901 pKa = 11.84NGGPGVEE908 pKa = 4.04EE909 pKa = 5.07FYY911 pKa = 10.99LSSVEE916 pKa = 4.15EE917 pKa = 4.41TDD919 pKa = 6.09DD920 pKa = 4.26SDD922 pKa = 4.03PDD924 pKa = 3.66NPVTTRR930 pKa = 11.84CQEE933 pKa = 4.14CFDD936 pKa = 4.03SAVEE940 pKa = 3.82RR941 pKa = 11.84RR942 pKa = 11.84EE943 pKa = 3.74IEE945 pKa = 3.96QCILASRR952 pKa = 11.84AAISQTGGLPLGALQVGPDD971 pKa = 3.56GQIYY975 pKa = 9.1VAVVGSNQIGQILPGRR991 pKa = 11.84GCNPSTFNQDD1001 pKa = 2.41AVEE1004 pKa = 4.11PMPGTTNLGLPSFVQQSGSNIPEE1027 pKa = 4.2PSLAGPNRR1035 pKa = 11.84LCVGADD1041 pKa = 3.34EE1042 pKa = 4.79GAVGLFEE1049 pKa = 5.87GGGEE1053 pKa = 4.09PDD1055 pKa = 2.89IDD1057 pKa = 4.14LYY1059 pKa = 11.62NWTIFNEE1066 pKa = 3.94QGEE1069 pKa = 4.55IVDD1072 pKa = 3.61QFLNGAEE1079 pKa = 3.9EE1080 pKa = 4.18LQNLEE1085 pKa = 4.16YY1086 pKa = 10.91QFATAGLFTVQLEE1099 pKa = 4.4VNRR1102 pKa = 11.84CGEE1105 pKa = 3.81PWDD1108 pKa = 3.98EE1109 pKa = 4.08IFRR1112 pKa = 11.84LEE1114 pKa = 4.2VEE1116 pKa = 4.71VVDD1119 pKa = 4.16SPEE1122 pKa = 5.24IILPTDD1128 pKa = 2.99IALCADD1134 pKa = 3.58AVLEE1138 pKa = 4.26LVAVDD1143 pKa = 4.25PDD1145 pKa = 3.84DD1146 pKa = 4.38PRR1148 pKa = 11.84LDD1150 pKa = 3.5EE1151 pKa = 5.85YY1152 pKa = 11.6VFEE1155 pKa = 4.46WVNAAGEE1162 pKa = 4.32IVGDD1166 pKa = 3.83QNVLEE1171 pKa = 4.27VTEE1174 pKa = 4.07EE1175 pKa = 3.96SIYY1178 pKa = 10.55TVTVAYY1184 pKa = 9.89RR1185 pKa = 11.84LPEE1188 pKa = 4.45DD1189 pKa = 4.62ADD1191 pKa = 3.42PEE1193 pKa = 4.3MFQTCPVSRR1202 pKa = 11.84SVFVGPPFDD1211 pKa = 4.63FDD1213 pKa = 3.43IDD1215 pKa = 3.62QSEE1218 pKa = 4.53EE1219 pKa = 4.2EE1220 pKa = 4.42VCLGEE1225 pKa = 4.41TVTFTPNSPVSGSWSIHH1242 pKa = 5.92RR1243 pKa = 11.84EE1244 pKa = 3.8GSQDD1248 pKa = 4.1RR1249 pKa = 11.84ISLPDD1254 pKa = 3.66TLQLTLNTAILGEE1267 pKa = 4.38AGDD1270 pKa = 4.02YY1271 pKa = 11.14EE1272 pKa = 5.6LFFLTSDD1279 pKa = 4.33PLDD1282 pKa = 3.64EE1283 pKa = 4.46TCLFEE1288 pKa = 5.45RR1289 pKa = 11.84SAFLRR1294 pKa = 11.84VNAGPQFEE1302 pKa = 5.06VIATSPAQSCEE1313 pKa = 3.65IGDD1316 pKa = 3.99GVLEE1320 pKa = 4.13IQAGVLLDD1328 pKa = 3.6SLVVQEE1334 pKa = 4.32TGQLFFDD1341 pKa = 4.36VLPNDD1346 pKa = 4.19IIVVDD1351 pKa = 4.08GLEE1354 pKa = 4.06PGIYY1358 pKa = 9.45TLLGFANGCSSSQAAVIEE1376 pKa = 4.3NANPPSDD1383 pKa = 3.51VLFEE1387 pKa = 4.42LEE1389 pKa = 4.75TRR1391 pKa = 11.84DD1392 pKa = 3.74EE1393 pKa = 4.14QCAPEE1398 pKa = 4.24GLVAGGISILFGNGPASGVYY1418 pKa = 9.31EE1419 pKa = 4.17VLGLDD1424 pKa = 3.24TGEE1427 pKa = 5.05LISEE1431 pKa = 4.36SFANQSVIALDD1442 pKa = 4.23LPPGRR1447 pKa = 11.84YY1448 pKa = 8.73LVSVTDD1454 pKa = 3.44VGGCEE1459 pKa = 3.85VPAPSEE1465 pKa = 4.18VIVNEE1470 pKa = 3.9FSEE1473 pKa = 4.59SVEE1476 pKa = 4.05VEE1478 pKa = 3.97LFSPNLCGPVVSTSITASGDD1498 pKa = 3.16FLAVDD1503 pKa = 3.88RR1504 pKa = 11.84FGWFRR1509 pKa = 11.84EE1510 pKa = 4.12VNGEE1514 pKa = 4.35FIPITGEE1521 pKa = 3.6SGMVISVSEE1530 pKa = 3.94PGRR1533 pKa = 11.84YY1534 pKa = 7.73EE1535 pKa = 3.54VRR1537 pKa = 11.84LYY1539 pKa = 11.47NEE1541 pKa = 4.49FDD1543 pKa = 4.31CIIGSGVIDD1552 pKa = 4.45VIQSATEE1559 pKa = 3.88PPVLSPRR1566 pKa = 11.84YY1567 pKa = 9.31VICTLDD1573 pKa = 3.45GNLAEE1578 pKa = 4.57LQAGAGFASYY1588 pKa = 10.1SWSLDD1593 pKa = 3.16GQEE1596 pKa = 4.77LGRR1599 pKa = 11.84EE1600 pKa = 4.14ARR1602 pKa = 11.84FVPTLPGRR1610 pKa = 11.84YY1611 pKa = 8.48EE1612 pKa = 4.04LLVTDD1617 pKa = 3.95EE1618 pKa = 4.94AGCEE1622 pKa = 3.8FLVDD1626 pKa = 4.36FEE1628 pKa = 4.76VEE1630 pKa = 4.0EE1631 pKa = 4.39DD1632 pKa = 3.73CEE1634 pKa = 4.63INVRR1638 pKa = 11.84FPNAIIPGSPTRR1650 pKa = 11.84NFLLYY1655 pKa = 10.24TRR1657 pKa = 11.84GPIDD1661 pKa = 3.23KK1662 pKa = 10.29LGVYY1666 pKa = 9.74IYY1668 pKa = 10.62NRR1670 pKa = 11.84WGEE1673 pKa = 4.05LIYY1676 pKa = 11.0YY1677 pKa = 10.01CEE1679 pKa = 4.01EE1680 pKa = 4.15TNPSEE1685 pKa = 4.55EE1686 pKa = 4.37ISICTWDD1693 pKa = 4.26GYY1695 pKa = 11.74VNGQKK1700 pKa = 10.13VPVGTYY1706 pKa = 8.52PVVVKK1711 pKa = 9.01FSNQQQQIEE1720 pKa = 4.32RR1721 pKa = 11.84TIRR1724 pKa = 11.84RR1725 pKa = 11.84AIVVIEE1731 pKa = 3.9
Molecular weight: 187.44 kDa
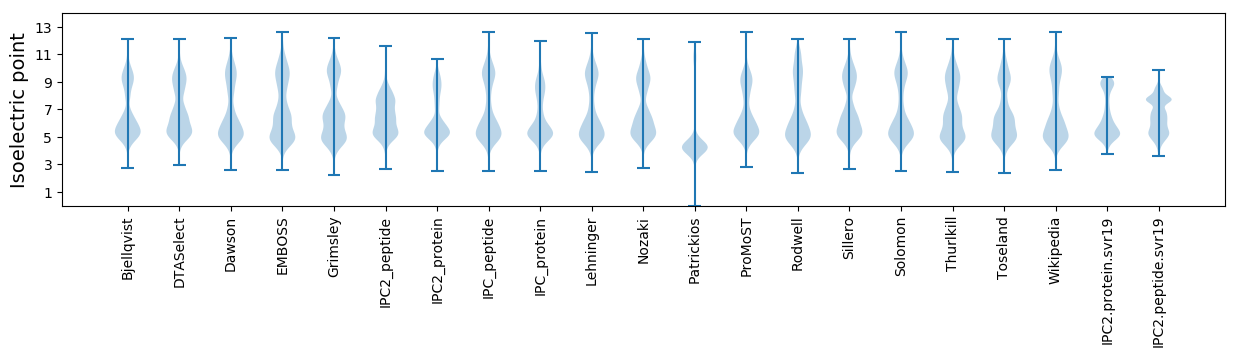
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7ZTC9|R7ZTC9_9BACT Molybdenum transport system permease OS=Lunatimonas lonarensis OX=1232681 GN=ADIS_2253 PE=3 SV=1
MM1 pKa = 7.15GRR3 pKa = 11.84EE4 pKa = 3.77KK5 pKa = 10.75AHH7 pKa = 6.57RR8 pKa = 11.84SAIIVARR15 pKa = 11.84GAAQPLVKK23 pKa = 10.02NAHH26 pKa = 5.64FRR28 pKa = 11.84WVLAAWTVEE37 pKa = 3.97EE38 pKa = 4.86ASLFRR43 pKa = 11.84QNPVLFGSALSRR55 pKa = 11.84CLGPGTDD62 pKa = 4.29VSPSSQGFWRR72 pKa = 11.84EE73 pKa = 3.85LVRR76 pKa = 11.84GIAMVAAKK84 pKa = 9.3TYY86 pKa = 10.16LSEE89 pKa = 4.47VVVKK93 pKa = 9.96PRR95 pKa = 11.84VEE97 pKa = 4.02TRR99 pKa = 11.84SYY101 pKa = 9.96PGWVV105 pKa = 3.11
MM1 pKa = 7.15GRR3 pKa = 11.84EE4 pKa = 3.77KK5 pKa = 10.75AHH7 pKa = 6.57RR8 pKa = 11.84SAIIVARR15 pKa = 11.84GAAQPLVKK23 pKa = 10.02NAHH26 pKa = 5.64FRR28 pKa = 11.84WVLAAWTVEE37 pKa = 3.97EE38 pKa = 4.86ASLFRR43 pKa = 11.84QNPVLFGSALSRR55 pKa = 11.84CLGPGTDD62 pKa = 4.29VSPSSQGFWRR72 pKa = 11.84EE73 pKa = 3.85LVRR76 pKa = 11.84GIAMVAAKK84 pKa = 9.3TYY86 pKa = 10.16LSEE89 pKa = 4.47VVVKK93 pKa = 9.96PRR95 pKa = 11.84VEE97 pKa = 4.02TRR99 pKa = 11.84SYY101 pKa = 9.96PGWVV105 pKa = 3.11
Molecular weight: 11.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1591650 |
37 |
2922 |
329.9 |
37.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.086 ± 0.037 | 0.704 ± 0.011 |
5.446 ± 0.025 | 6.595 ± 0.036 |
5.204 ± 0.03 | 7.487 ± 0.035 |
2.048 ± 0.019 | 6.509 ± 0.03 |
5.62 ± 0.041 | 9.982 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.427 ± 0.016 | 4.6 ± 0.035 |
4.325 ± 0.023 | 3.678 ± 0.017 |
5.07 ± 0.023 | 6.389 ± 0.025 |
5.081 ± 0.024 | 6.661 ± 0.027 |
1.382 ± 0.018 | 3.706 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
