
Verrucomicrobiaceae bacterium SCGC AG-212-N21
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Verrucomicrobiae; Verrucomicrobiales; Verrucomicrobiaceae; unclassified Verrucomicrobiaceae
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
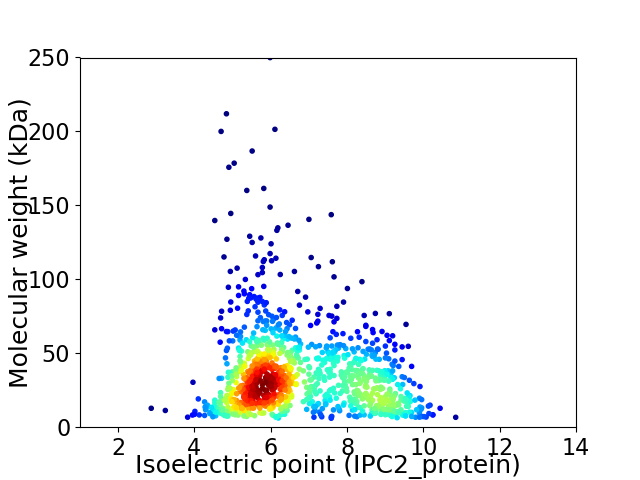
Virtual 2D-PAGE plot for 1135 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177RF53|A0A177RF53_9BACT SHSP domain-containing protein OS=Verrucomicrobiaceae bacterium SCGC AG-212-N21 OX=1799659 GN=AYO49_06305 PE=3 SV=1
MM1 pKa = 7.59KK2 pKa = 10.59LILHH6 pKa = 6.86LLFASALITSASAGTYY22 pKa = 8.4TSSKK26 pKa = 8.71SAKK29 pKa = 9.84AVVPPAPLGCDD40 pKa = 3.18CFAPGFAIGIFGSGVLFNEE59 pKa = 5.07DD60 pKa = 5.49DD61 pKa = 4.07IDD63 pKa = 5.19DD64 pKa = 4.75DD65 pKa = 4.38SLGGGVLAEE74 pKa = 4.11YY75 pKa = 10.31FFTEE79 pKa = 4.25NIGIQGSYY87 pKa = 11.09GIFATEE93 pKa = 4.13KK94 pKa = 9.74EE95 pKa = 4.24LHH97 pKa = 6.48EE98 pKa = 4.51FDD100 pKa = 3.57AALVLRR106 pKa = 11.84YY107 pKa = 9.11PITSLCLAPYY117 pKa = 10.25IMGGAGYY124 pKa = 8.03STNSQDD130 pKa = 3.47AWNLFLGGGIDD141 pKa = 3.83VRR143 pKa = 11.84CPEE146 pKa = 4.41WNCVSLFADD155 pKa = 5.06GAYY158 pKa = 9.7HH159 pKa = 7.18WSEE162 pKa = 5.23DD163 pKa = 3.65DD164 pKa = 4.7DD165 pKa = 6.44ADD167 pKa = 3.8FTIVRR172 pKa = 11.84IGLKK176 pKa = 10.38FPLL179 pKa = 4.39
MM1 pKa = 7.59KK2 pKa = 10.59LILHH6 pKa = 6.86LLFASALITSASAGTYY22 pKa = 8.4TSSKK26 pKa = 8.71SAKK29 pKa = 9.84AVVPPAPLGCDD40 pKa = 3.18CFAPGFAIGIFGSGVLFNEE59 pKa = 5.07DD60 pKa = 5.49DD61 pKa = 4.07IDD63 pKa = 5.19DD64 pKa = 4.75DD65 pKa = 4.38SLGGGVLAEE74 pKa = 4.11YY75 pKa = 10.31FFTEE79 pKa = 4.25NIGIQGSYY87 pKa = 11.09GIFATEE93 pKa = 4.13KK94 pKa = 9.74EE95 pKa = 4.24LHH97 pKa = 6.48EE98 pKa = 4.51FDD100 pKa = 3.57AALVLRR106 pKa = 11.84YY107 pKa = 9.11PITSLCLAPYY117 pKa = 10.25IMGGAGYY124 pKa = 8.03STNSQDD130 pKa = 3.47AWNLFLGGGIDD141 pKa = 3.83VRR143 pKa = 11.84CPEE146 pKa = 4.41WNCVSLFADD155 pKa = 5.06GAYY158 pKa = 9.7HH159 pKa = 7.18WSEE162 pKa = 5.23DD163 pKa = 3.65DD164 pKa = 4.7DD165 pKa = 6.44ADD167 pKa = 3.8FTIVRR172 pKa = 11.84IGLKK176 pKa = 10.38FPLL179 pKa = 4.39
Molecular weight: 19.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177RAJ0|A0A177RAJ0_9BACT Uncharacterized protein OS=Verrucomicrobiaceae bacterium SCGC AG-212-N21 OX=1799659 GN=AYO49_04755 PE=4 SV=1
MM1 pKa = 7.76PMQHH5 pKa = 6.35EE6 pKa = 5.21AIQLEE11 pKa = 4.42AGQSFRR17 pKa = 11.84LLHH20 pKa = 6.12WKK22 pKa = 10.51DD23 pKa = 3.3NLRR26 pKa = 11.84EE27 pKa = 3.92VDD29 pKa = 3.76SLIGPEE35 pKa = 3.88EE36 pKa = 3.94RR37 pKa = 11.84VRR39 pKa = 11.84IHH41 pKa = 6.38GAGDD45 pKa = 3.06RR46 pKa = 11.84WHH48 pKa = 6.84HH49 pKa = 5.47HH50 pKa = 6.46RR51 pKa = 11.84EE52 pKa = 3.78MEE54 pKa = 4.1LTVVQRR60 pKa = 11.84GSGTRR65 pKa = 11.84FVGDD69 pKa = 3.73HH70 pKa = 6.64IGPFEE75 pKa = 4.28SLDD78 pKa = 3.59VVLIGANVPHH88 pKa = 6.97FWKK91 pKa = 10.66GMRR94 pKa = 11.84KK95 pKa = 8.71SSGYY99 pKa = 8.5AVQWRR104 pKa = 11.84FEE106 pKa = 4.18RR107 pKa = 11.84EE108 pKa = 3.41HH109 pKa = 7.3ALWSFPEE116 pKa = 4.22SGALRR121 pKa = 11.84VLWEE125 pKa = 4.11RR126 pKa = 11.84AAHH129 pKa = 6.02GLRR132 pKa = 11.84FTGATADD139 pKa = 3.77RR140 pKa = 11.84ALALVQSMARR150 pKa = 11.84RR151 pKa = 11.84EE152 pKa = 4.42GCGRR156 pKa = 11.84LAMLFEE162 pKa = 5.24LLDD165 pKa = 4.01KK166 pKa = 10.95LATAPRR172 pKa = 11.84RR173 pKa = 11.84EE174 pKa = 4.18CQKK177 pKa = 10.65LSQRR181 pKa = 11.84PFDD184 pKa = 3.9LSGAHH189 pKa = 5.95AHH191 pKa = 5.62QPAIEE196 pKa = 3.98RR197 pKa = 11.84VIRR200 pKa = 11.84YY201 pKa = 8.57ILQHH205 pKa = 5.5FRR207 pKa = 11.84EE208 pKa = 5.15PIALAEE214 pKa = 4.16VLRR217 pKa = 11.84LAGMSKK223 pKa = 9.24ATFARR228 pKa = 11.84QFRR231 pKa = 11.84RR232 pKa = 11.84HH233 pKa = 5.87AGRR236 pKa = 11.84PFSAFVNQVRR246 pKa = 11.84LDD248 pKa = 3.57HH249 pKa = 6.84ACRR252 pKa = 11.84EE253 pKa = 4.34LVGSGMSVSEE263 pKa = 4.03AAFASGFNSLSYY275 pKa = 10.34FNRR278 pKa = 11.84AFRR281 pKa = 11.84SAHH284 pKa = 5.16RR285 pKa = 11.84CSPKK289 pKa = 9.68EE290 pKa = 3.72YY291 pKa = 9.49RR292 pKa = 11.84RR293 pKa = 11.84KK294 pKa = 10.49ARR296 pKa = 11.84VSLL299 pKa = 3.86
MM1 pKa = 7.76PMQHH5 pKa = 6.35EE6 pKa = 5.21AIQLEE11 pKa = 4.42AGQSFRR17 pKa = 11.84LLHH20 pKa = 6.12WKK22 pKa = 10.51DD23 pKa = 3.3NLRR26 pKa = 11.84EE27 pKa = 3.92VDD29 pKa = 3.76SLIGPEE35 pKa = 3.88EE36 pKa = 3.94RR37 pKa = 11.84VRR39 pKa = 11.84IHH41 pKa = 6.38GAGDD45 pKa = 3.06RR46 pKa = 11.84WHH48 pKa = 6.84HH49 pKa = 5.47HH50 pKa = 6.46RR51 pKa = 11.84EE52 pKa = 3.78MEE54 pKa = 4.1LTVVQRR60 pKa = 11.84GSGTRR65 pKa = 11.84FVGDD69 pKa = 3.73HH70 pKa = 6.64IGPFEE75 pKa = 4.28SLDD78 pKa = 3.59VVLIGANVPHH88 pKa = 6.97FWKK91 pKa = 10.66GMRR94 pKa = 11.84KK95 pKa = 8.71SSGYY99 pKa = 8.5AVQWRR104 pKa = 11.84FEE106 pKa = 4.18RR107 pKa = 11.84EE108 pKa = 3.41HH109 pKa = 7.3ALWSFPEE116 pKa = 4.22SGALRR121 pKa = 11.84VLWEE125 pKa = 4.11RR126 pKa = 11.84AAHH129 pKa = 6.02GLRR132 pKa = 11.84FTGATADD139 pKa = 3.77RR140 pKa = 11.84ALALVQSMARR150 pKa = 11.84RR151 pKa = 11.84EE152 pKa = 4.42GCGRR156 pKa = 11.84LAMLFEE162 pKa = 5.24LLDD165 pKa = 4.01KK166 pKa = 10.95LATAPRR172 pKa = 11.84RR173 pKa = 11.84EE174 pKa = 4.18CQKK177 pKa = 10.65LSQRR181 pKa = 11.84PFDD184 pKa = 3.9LSGAHH189 pKa = 5.95AHH191 pKa = 5.62QPAIEE196 pKa = 3.98RR197 pKa = 11.84VIRR200 pKa = 11.84YY201 pKa = 8.57ILQHH205 pKa = 5.5FRR207 pKa = 11.84EE208 pKa = 5.15PIALAEE214 pKa = 4.16VLRR217 pKa = 11.84LAGMSKK223 pKa = 9.24ATFARR228 pKa = 11.84QFRR231 pKa = 11.84RR232 pKa = 11.84HH233 pKa = 5.87AGRR236 pKa = 11.84PFSAFVNQVRR246 pKa = 11.84LDD248 pKa = 3.57HH249 pKa = 6.84ACRR252 pKa = 11.84EE253 pKa = 4.34LVGSGMSVSEE263 pKa = 4.03AAFASGFNSLSYY275 pKa = 10.34FNRR278 pKa = 11.84AFRR281 pKa = 11.84SAHH284 pKa = 5.16RR285 pKa = 11.84CSPKK289 pKa = 9.68EE290 pKa = 3.72YY291 pKa = 9.49RR292 pKa = 11.84RR293 pKa = 11.84KK294 pKa = 10.49ARR296 pKa = 11.84VSLL299 pKa = 3.86
Molecular weight: 34.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
384530 |
57 |
2303 |
338.8 |
37.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.721 ± 0.086 | 1.068 ± 0.023 |
5.424 ± 0.052 | 5.886 ± 0.066 |
3.947 ± 0.045 | 8.143 ± 0.094 |
2.411 ± 0.038 | 4.442 ± 0.047 |
4.556 ± 0.072 | 9.85 ± 0.087 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.377 ± 0.037 | 3.066 ± 0.059 |
5.481 ± 0.061 | 3.427 ± 0.045 |
6.329 ± 0.07 | 5.845 ± 0.068 |
5.74 ± 0.085 | 7.199 ± 0.062 |
1.702 ± 0.035 | 2.385 ± 0.045 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
