
Fusarium venenatum
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Nectriaceae; Fusarium; Fusarium sambucinum species complex
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
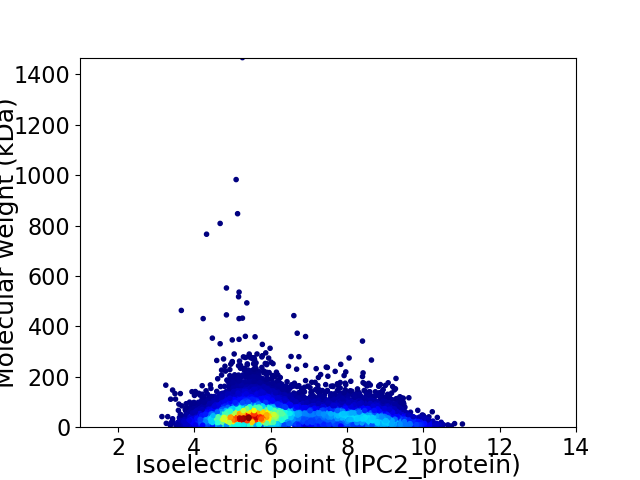
Virtual 2D-PAGE plot for 13945 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2L2T2E5|A0A2L2T2E5_9HYPO Uncharacterized protein OS=Fusarium venenatum OX=56646 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 10.58AFIFIAALLTAAEE15 pKa = 4.18AVLAKK20 pKa = 10.71DD21 pKa = 3.99LVAVEE26 pKa = 4.9APADD30 pKa = 3.82TWCITYY36 pKa = 10.58LSTYY40 pKa = 9.64LATVTNPGDD49 pKa = 3.9ILPSDD54 pKa = 3.58ASGNDD59 pKa = 3.58GPDD62 pKa = 3.23AQNSGRR68 pKa = 11.84LPFAPSLHH76 pKa = 5.5PTFRR80 pKa = 11.84RR81 pKa = 11.84NTSISVTRR89 pKa = 11.84SAMSTDD95 pKa = 3.15VRR97 pKa = 11.84DD98 pKa = 3.74TLGPEE103 pKa = 4.14SSLSTSVVINTVDD116 pKa = 3.36TSPVNSEE123 pKa = 4.13SSDD126 pKa = 3.81LDD128 pKa = 3.47VDD130 pKa = 3.85PTAAQSSYY138 pKa = 10.89GVSTITDD145 pKa = 3.86GLSTMAASTVDD156 pKa = 3.65AEE158 pKa = 4.5ATSTSTDD165 pKa = 3.07IIEE168 pKa = 4.13PAGRR172 pKa = 11.84IVIFLIQTTDD182 pKa = 2.99NEE184 pKa = 3.57KK185 pKa = 9.17RR186 pKa = 11.84AIYY189 pKa = 9.96RR190 pKa = 11.84RR191 pKa = 11.84VISGFIGLNNPSVCTFAATFNLAEE215 pKa = 4.45GQLFADD221 pKa = 5.2GVPIYY226 pKa = 11.02YY227 pKa = 10.32SGEE230 pKa = 4.04DD231 pKa = 3.68YY232 pKa = 11.25KK233 pKa = 11.19EE234 pKa = 4.47LSAQGRR240 pKa = 11.84PPSGSITSGFADD252 pKa = 3.35SGGTLTFRR260 pKa = 11.84NSDD263 pKa = 3.98LPNGEE268 pKa = 5.23AGFCQDD274 pKa = 4.7ADD276 pKa = 3.72GQVYY280 pKa = 7.83ITFTTGPSGCVPVNLAAYY298 pKa = 9.83NVEE301 pKa = 3.9QCQDD305 pKa = 2.84GRR307 pKa = 11.84LIGLDD312 pKa = 3.56EE313 pKa = 4.4LTSSVSGTAITTADD327 pKa = 3.43QSFSAPSTFGEE338 pKa = 4.47LPEE341 pKa = 4.4STDD344 pKa = 4.45LIQSQSIIPSSQTSEE359 pKa = 4.46FEE361 pKa = 3.94ASATSLSHH369 pKa = 4.91VTEE372 pKa = 4.67SSTQEE377 pKa = 3.67LGGASSQEE385 pKa = 3.97TSALTVRR392 pKa = 11.84EE393 pKa = 4.0DD394 pKa = 3.96LEE396 pKa = 4.52TLSSVLTTSGDD407 pKa = 3.8SASEE411 pKa = 4.07PTSASTPHH419 pKa = 6.48LSSEE423 pKa = 4.29TSDD426 pKa = 4.82LLDD429 pKa = 3.6STTTSSDD436 pKa = 2.92GTEE439 pKa = 3.99LSTTSGVEE447 pKa = 4.04SQTTTGIEE455 pKa = 4.02TSHH458 pKa = 7.85IEE460 pKa = 4.17TTIKK464 pKa = 10.09TSVSEE469 pKa = 3.98TTIILEE475 pKa = 4.34TTEE478 pKa = 4.03ATPTEE483 pKa = 4.56GDD485 pKa = 3.34TSSTEE490 pKa = 4.0VVSDD494 pKa = 3.47VDD496 pKa = 3.64TTAATDD502 pKa = 3.68TTDD505 pKa = 3.26TTTTNADD512 pKa = 3.69VITDD516 pKa = 4.44LEE518 pKa = 4.83DD519 pKa = 3.46STTMEE524 pKa = 4.63AEE526 pKa = 4.01TTAAEE531 pKa = 4.48TTTSVKK537 pKa = 9.31PTTTTEE543 pKa = 4.05TATEE547 pKa = 4.31PEE549 pKa = 4.42DD550 pKa = 3.59LTTTEE555 pKa = 5.2AEE557 pKa = 4.32TTTPEE562 pKa = 3.61ITTTAEE568 pKa = 3.83LTTTTEE574 pKa = 4.22AAPEE578 pKa = 4.06PAPQFACGDD587 pKa = 3.71PGYY590 pKa = 7.01TTTYY594 pKa = 9.01TYY596 pKa = 11.62NSVTFNLQCEE606 pKa = 4.51SNYY609 pKa = 10.71SYY611 pKa = 11.24FGLEE615 pKa = 4.18TFSTASFGEE624 pKa = 4.33CLSRR628 pKa = 11.84CALNSACNGIVWRR641 pKa = 11.84RR642 pKa = 11.84ASLLCSLTTGVTAYY656 pKa = 10.53GPDD659 pKa = 3.12NRR661 pKa = 11.84FDD663 pKa = 3.61VATVASRR670 pKa = 11.84AA671 pKa = 3.56
MM1 pKa = 7.47KK2 pKa = 10.58AFIFIAALLTAAEE15 pKa = 4.18AVLAKK20 pKa = 10.71DD21 pKa = 3.99LVAVEE26 pKa = 4.9APADD30 pKa = 3.82TWCITYY36 pKa = 10.58LSTYY40 pKa = 9.64LATVTNPGDD49 pKa = 3.9ILPSDD54 pKa = 3.58ASGNDD59 pKa = 3.58GPDD62 pKa = 3.23AQNSGRR68 pKa = 11.84LPFAPSLHH76 pKa = 5.5PTFRR80 pKa = 11.84RR81 pKa = 11.84NTSISVTRR89 pKa = 11.84SAMSTDD95 pKa = 3.15VRR97 pKa = 11.84DD98 pKa = 3.74TLGPEE103 pKa = 4.14SSLSTSVVINTVDD116 pKa = 3.36TSPVNSEE123 pKa = 4.13SSDD126 pKa = 3.81LDD128 pKa = 3.47VDD130 pKa = 3.85PTAAQSSYY138 pKa = 10.89GVSTITDD145 pKa = 3.86GLSTMAASTVDD156 pKa = 3.65AEE158 pKa = 4.5ATSTSTDD165 pKa = 3.07IIEE168 pKa = 4.13PAGRR172 pKa = 11.84IVIFLIQTTDD182 pKa = 2.99NEE184 pKa = 3.57KK185 pKa = 9.17RR186 pKa = 11.84AIYY189 pKa = 9.96RR190 pKa = 11.84RR191 pKa = 11.84VISGFIGLNNPSVCTFAATFNLAEE215 pKa = 4.45GQLFADD221 pKa = 5.2GVPIYY226 pKa = 11.02YY227 pKa = 10.32SGEE230 pKa = 4.04DD231 pKa = 3.68YY232 pKa = 11.25KK233 pKa = 11.19EE234 pKa = 4.47LSAQGRR240 pKa = 11.84PPSGSITSGFADD252 pKa = 3.35SGGTLTFRR260 pKa = 11.84NSDD263 pKa = 3.98LPNGEE268 pKa = 5.23AGFCQDD274 pKa = 4.7ADD276 pKa = 3.72GQVYY280 pKa = 7.83ITFTTGPSGCVPVNLAAYY298 pKa = 9.83NVEE301 pKa = 3.9QCQDD305 pKa = 2.84GRR307 pKa = 11.84LIGLDD312 pKa = 3.56EE313 pKa = 4.4LTSSVSGTAITTADD327 pKa = 3.43QSFSAPSTFGEE338 pKa = 4.47LPEE341 pKa = 4.4STDD344 pKa = 4.45LIQSQSIIPSSQTSEE359 pKa = 4.46FEE361 pKa = 3.94ASATSLSHH369 pKa = 4.91VTEE372 pKa = 4.67SSTQEE377 pKa = 3.67LGGASSQEE385 pKa = 3.97TSALTVRR392 pKa = 11.84EE393 pKa = 4.0DD394 pKa = 3.96LEE396 pKa = 4.52TLSSVLTTSGDD407 pKa = 3.8SASEE411 pKa = 4.07PTSASTPHH419 pKa = 6.48LSSEE423 pKa = 4.29TSDD426 pKa = 4.82LLDD429 pKa = 3.6STTTSSDD436 pKa = 2.92GTEE439 pKa = 3.99LSTTSGVEE447 pKa = 4.04SQTTTGIEE455 pKa = 4.02TSHH458 pKa = 7.85IEE460 pKa = 4.17TTIKK464 pKa = 10.09TSVSEE469 pKa = 3.98TTIILEE475 pKa = 4.34TTEE478 pKa = 4.03ATPTEE483 pKa = 4.56GDD485 pKa = 3.34TSSTEE490 pKa = 4.0VVSDD494 pKa = 3.47VDD496 pKa = 3.64TTAATDD502 pKa = 3.68TTDD505 pKa = 3.26TTTTNADD512 pKa = 3.69VITDD516 pKa = 4.44LEE518 pKa = 4.83DD519 pKa = 3.46STTMEE524 pKa = 4.63AEE526 pKa = 4.01TTAAEE531 pKa = 4.48TTTSVKK537 pKa = 9.31PTTTTEE543 pKa = 4.05TATEE547 pKa = 4.31PEE549 pKa = 4.42DD550 pKa = 3.59LTTTEE555 pKa = 5.2AEE557 pKa = 4.32TTTPEE562 pKa = 3.61ITTTAEE568 pKa = 3.83LTTTTEE574 pKa = 4.22AAPEE578 pKa = 4.06PAPQFACGDD587 pKa = 3.71PGYY590 pKa = 7.01TTTYY594 pKa = 9.01TYY596 pKa = 11.62NSVTFNLQCEE606 pKa = 4.51SNYY609 pKa = 10.71SYY611 pKa = 11.24FGLEE615 pKa = 4.18TFSTASFGEE624 pKa = 4.33CLSRR628 pKa = 11.84CALNSACNGIVWRR641 pKa = 11.84RR642 pKa = 11.84ASLLCSLTTGVTAYY656 pKa = 10.53GPDD659 pKa = 3.12NRR661 pKa = 11.84FDD663 pKa = 3.61VATVASRR670 pKa = 11.84AA671 pKa = 3.56
Molecular weight: 69.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2L2TAM3|A0A2L2TAM3_9HYPO NAD_binding_2 domain-containing protein OS=Fusarium venenatum OX=56646 PE=4 SV=1
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.34RR8 pKa = 11.84HH9 pKa = 4.3SAPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APARR26 pKa = 11.84SNRR29 pKa = 11.84HH30 pKa = 4.32TVTTTTTTTTKK41 pKa = 9.91PRR43 pKa = 11.84RR44 pKa = 11.84GMFGGGSTARR54 pKa = 11.84RR55 pKa = 11.84THH57 pKa = 5.96GAAPVHH63 pKa = 5.35HH64 pKa = 6.52HH65 pKa = 5.47QRR67 pKa = 11.84RR68 pKa = 11.84PSMKK72 pKa = 10.02DD73 pKa = 2.95KK74 pKa = 11.41VSGALLKK81 pKa = 10.61IKK83 pKa = 10.52GSLTRR88 pKa = 11.84RR89 pKa = 11.84PGVKK93 pKa = 9.89AAGTRR98 pKa = 11.84RR99 pKa = 11.84MHH101 pKa = 5.65GTDD104 pKa = 2.95GRR106 pKa = 11.84GARR109 pKa = 11.84HH110 pKa = 5.73RR111 pKa = 11.84RR112 pKa = 11.84YY113 pKa = 10.37
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.34RR8 pKa = 11.84HH9 pKa = 4.3SAPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APARR26 pKa = 11.84SNRR29 pKa = 11.84HH30 pKa = 4.32TVTTTTTTTTKK41 pKa = 9.91PRR43 pKa = 11.84RR44 pKa = 11.84GMFGGGSTARR54 pKa = 11.84RR55 pKa = 11.84THH57 pKa = 5.96GAAPVHH63 pKa = 5.35HH64 pKa = 6.52HH65 pKa = 5.47QRR67 pKa = 11.84RR68 pKa = 11.84PSMKK72 pKa = 10.02DD73 pKa = 2.95KK74 pKa = 11.41VSGALLKK81 pKa = 10.61IKK83 pKa = 10.52GSLTRR88 pKa = 11.84RR89 pKa = 11.84PGVKK93 pKa = 9.89AAGTRR98 pKa = 11.84RR99 pKa = 11.84MHH101 pKa = 5.65GTDD104 pKa = 2.95GRR106 pKa = 11.84GARR109 pKa = 11.84HH110 pKa = 5.73RR111 pKa = 11.84RR112 pKa = 11.84YY113 pKa = 10.37
Molecular weight: 12.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6439375 |
10 |
13331 |
461.8 |
51.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.183 ± 0.018 | 1.314 ± 0.008 |
5.881 ± 0.017 | 6.191 ± 0.024 |
3.783 ± 0.013 | 6.735 ± 0.02 |
2.365 ± 0.011 | 5.153 ± 0.016 |
5.077 ± 0.019 | 8.715 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.267 ± 0.007 | 3.883 ± 0.01 |
5.876 ± 0.024 | 4.032 ± 0.014 |
5.776 ± 0.021 | 8.167 ± 0.025 |
6.128 ± 0.026 | 6.136 ± 0.016 |
1.522 ± 0.009 | 2.817 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
